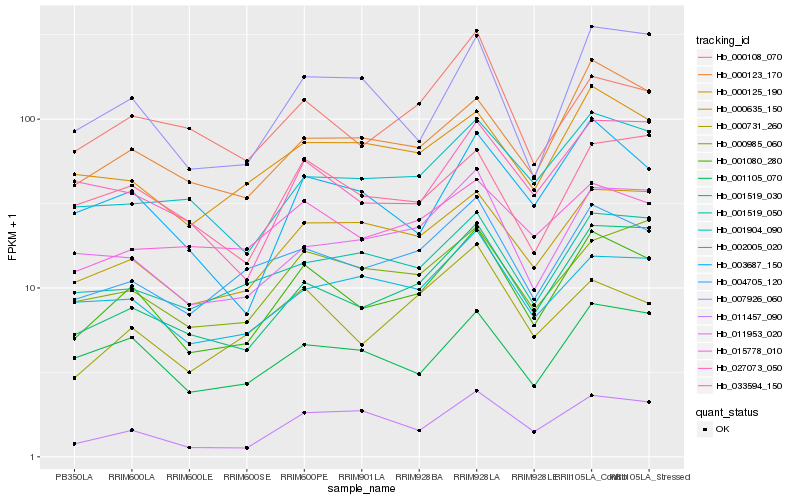
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 2 |
Hb_004705_120 |
0.0935721847 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001519_030 |
0.0938541522 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000635_150 |
0.1008381385 |
- |
- |
mitochondrial import receptor subunit TOM20-2 family protein [Populus trichocarpa] |
| 5 |
Hb_000123_170 |
0.1097909743 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 6 |
Hb_001105_070 |
0.1098076361 |
- |
- |
PREDICTED: uncharacterized protein LOC105632829 [Jatropha curcas] |
| 7 |
Hb_011953_020 |
0.1111622939 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000731_260 |
0.1125835827 |
- |
- |
hypothetical protein RCOM_0905090 [Ricinus communis] |
| 9 |
Hb_000985_060 |
0.1136802852 |
- |
- |
PREDICTED: protein transport protein SFT2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000125_190 |
0.114584013 |
- |
- |
PREDICTED: F-box protein SKIP16 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003687_150 |
0.1149107011 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002005_020 |
0.1172433517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000108_070 |
0.1175291029 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 14 |
Hb_001904_090 |
0.1177849332 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 15 |
Hb_011457_090 |
0.1193853537 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001519_050 |
0.1195257703 |
- |
- |
PREDICTED: protein RMD5 homolog A-like [Jatropha curcas] |
| 17 |
Hb_033594_150 |
0.12069767 |
- |
- |
unknown [Lotus japonicus] |
| 18 |
Hb_027073_050 |
0.1208238541 |
- |
- |
PREDICTED: uncharacterized protein LOC105635813 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007926_060 |
0.1209992256 |
- |
- |
ubiquitin-conjugating enzyme E2 variant 1D [Arabidopsis thaliana] |
| 20 |
Hb_015778_010 |
0.1220116012 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 1 [Jatropha curcas] |