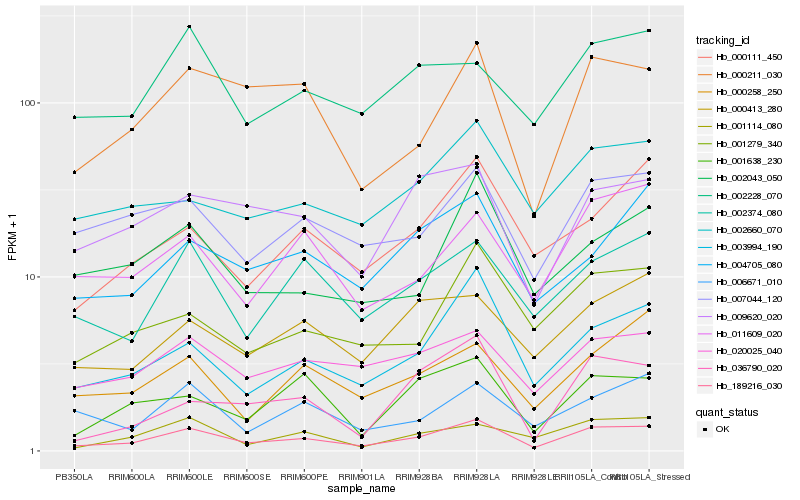
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_189216_030 |
0.0 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |
| 2 |
Hb_003994_190 |
0.1258173808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000211_030 |
0.1291318896 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 4 |
Hb_002374_080 |
0.1413293366 |
- |
- |
PREDICTED: chloroplast processing peptidase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007044_120 |
0.1463857579 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 6 |
Hb_009620_020 |
0.148123847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_020025_040 |
0.1490241357 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 8 |
Hb_011609_020 |
0.1505123937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001638_230 |
0.1515308016 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 10 |
Hb_001279_340 |
0.1516329717 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 11 |
Hb_000111_450 |
0.1531359908 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |
| 12 |
Hb_001114_080 |
0.1531583326 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 13 |
Hb_004705_080 |
0.1551810416 |
transcription factor |
TF Family: ARF |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002043_050 |
0.1553672875 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 15 |
Hb_006671_010 |
0.1554490264 |
- |
- |
PREDICTED: 5' exonuclease Apollo isoform X1 [Jatropha curcas] |
| 16 |
Hb_002660_070 |
0.1556944072 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 17 |
Hb_000413_280 |
0.1557536344 |
- |
- |
PREDICTED: probable UDP-3-O-acylglucosamine N-acyltransferase 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000258_250 |
0.1578286891 |
- |
- |
Origin recognition complex subunit, putative [Ricinus communis] |
| 19 |
Hb_002228_070 |
0.159719319 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 20 |
Hb_036790_020 |
0.1613044068 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |