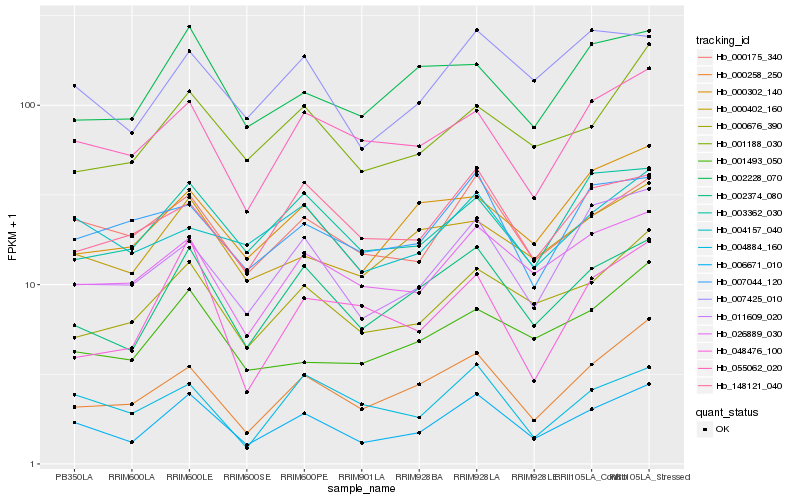
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006671_010 |
0.0 |
- |
- |
PREDICTED: 5' exonuclease Apollo isoform X1 [Jatropha curcas] |
| 2 |
Hb_002374_080 |
0.0847818712 |
- |
- |
PREDICTED: chloroplast processing peptidase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000175_340 |
0.1091637992 |
- |
- |
PREDICTED: uncharacterized protein LOC105634861 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000676_390 |
0.1110727593 |
- |
- |
PREDICTED: protein FMP32, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000258_250 |
0.1110842237 |
- |
- |
Origin recognition complex subunit, putative [Ricinus communis] |
| 6 |
Hb_011609_020 |
0.1112106234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_026889_030 |
0.1141489579 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_007425_010 |
0.1201300069 |
- |
- |
uncharacterized protein LOC100499741 [Glycine max] |
| 9 |
Hb_055062_020 |
0.1201328201 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 10 |
Hb_003362_030 |
0.1262335657 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 11 |
Hb_000402_160 |
0.1281362257 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001188_030 |
0.1294645874 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 13 |
Hb_001493_050 |
0.1310380022 |
- |
- |
PREDICTED: uncharacterized protein LOC105631312 [Jatropha curcas] |
| 14 |
Hb_004884_160 |
0.1339288066 |
- |
- |
PREDICTED: uncharacterized protein LOC105648105 [Jatropha curcas] |
| 15 |
Hb_148121_040 |
0.1355056144 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000302_140 |
0.1370829529 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 17 |
Hb_007044_120 |
0.1383930757 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 18 |
Hb_002228_070 |
0.1387417322 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 19 |
Hb_004157_040 |
0.1387500805 |
- |
- |
Rab2 [Hevea brasiliensis] |
| 20 |
Hb_048476_100 |
0.1414863116 |
- |
- |
PREDICTED: glutathione S-transferase U9-like [Vitis vinifera] |