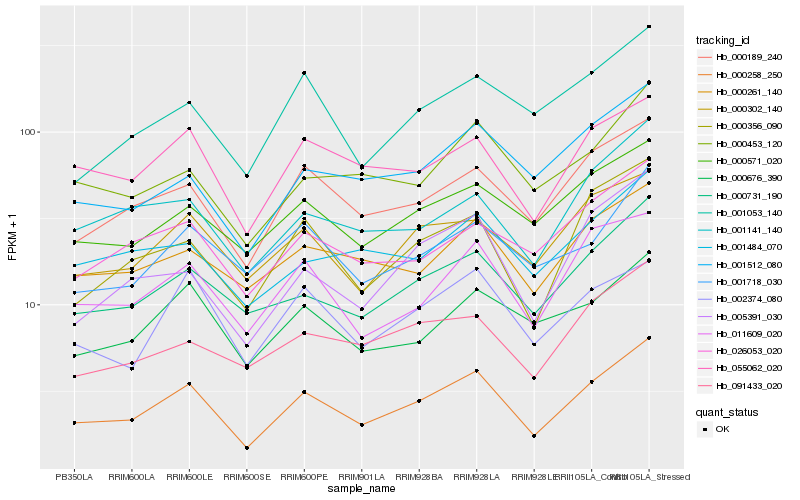
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_250 |
0.0 |
- |
- |
Origin recognition complex subunit, putative [Ricinus communis] |
| 2 |
Hb_000189_240 |
0.0875299097 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 3 |
Hb_000731_190 |
0.0921448171 |
- |
- |
PREDICTED: cardiolipin synthase (CMP-forming), mitochondrial [Jatropha curcas] |
| 4 |
Hb_000356_090 |
0.0927233096 |
- |
- |
PREDICTED: uncharacterized protein At2g38710 [Jatropha curcas] |
| 5 |
Hb_011609_020 |
0.0948980373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_055062_020 |
0.1000224885 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 7 |
Hb_000302_140 |
0.1025447161 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001141_140 |
0.1036045807 |
- |
- |
20S proteasome beta subunit D3 [Hevea brasiliensis] |
| 9 |
Hb_000571_020 |
0.1038868447 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005391_030 |
0.1047250446 |
- |
- |
putative cytidine/deoxycytidylate deaminase family protein [Zea mays] |
| 11 |
Hb_000261_140 |
0.1055097512 |
- |
- |
- |
| 12 |
Hb_001512_080 |
0.1055829745 |
- |
- |
PREDICTED: proteasome subunit beta type-7-B [Jatropha curcas] |
| 13 |
Hb_000453_120 |
0.1062848496 |
- |
- |
PREDICTED: proteasome subunit alpha type-3 [Jatropha curcas] |
| 14 |
Hb_026053_020 |
0.1075206005 |
- |
- |
PREDICTED: uncharacterized protein LOC105643718 [Jatropha curcas] |
| 15 |
Hb_002374_080 |
0.1083583261 |
- |
- |
PREDICTED: chloroplast processing peptidase-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_091433_020 |
0.1085983571 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000676_390 |
0.1086380978 |
- |
- |
PREDICTED: protein FMP32, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001718_030 |
0.1101479873 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001053_140 |
0.1102128488 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001484_070 |
0.1106398146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |