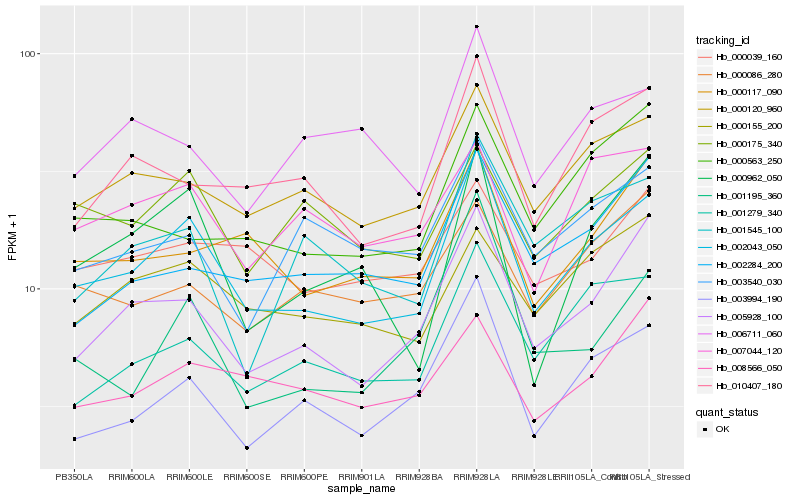
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002043_050 |
0.0 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 2 |
Hb_005928_100 |
0.0970988641 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2 [Jatropha curcas] |
| 3 |
Hb_000120_960 |
0.1048962464 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_003994_190 |
0.1121612632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001279_340 |
0.1145834383 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 6 |
Hb_010407_180 |
0.1153529538 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 7 |
Hb_000155_200 |
0.1217545972 |
- |
- |
Rab7 [Hevea brasiliensis] |
| 8 |
Hb_000117_090 |
0.1219039982 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 9 |
Hb_000962_050 |
0.1252812952 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 10 |
Hb_000175_340 |
0.1273667719 |
- |
- |
PREDICTED: uncharacterized protein LOC105634861 isoform X3 [Jatropha curcas] |
| 11 |
Hb_007044_120 |
0.1304297917 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 12 |
Hb_000039_160 |
0.1305792125 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC [Jatropha curcas] |
| 13 |
Hb_001545_100 |
0.1306188273 |
- |
- |
PREDICTED: probable F-actin-capping protein subunit beta [Vitis vinifera] |
| 14 |
Hb_008566_050 |
0.1318495035 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 15 |
Hb_000086_280 |
0.1329052751 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 3-like [Gossypium raimondii] |
| 16 |
Hb_002284_200 |
0.133699662 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006711_060 |
0.1338665379 |
- |
- |
Tubulin beta-1 chain [Morus notabilis] |
| 18 |
Hb_000563_250 |
0.1340319293 |
- |
- |
PREDICTED: uncharacterized protein LOC105643472 [Jatropha curcas] |
| 19 |
Hb_001195_360 |
0.1342076609 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 20 |
Hb_003540_030 |
0.1352420958 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |