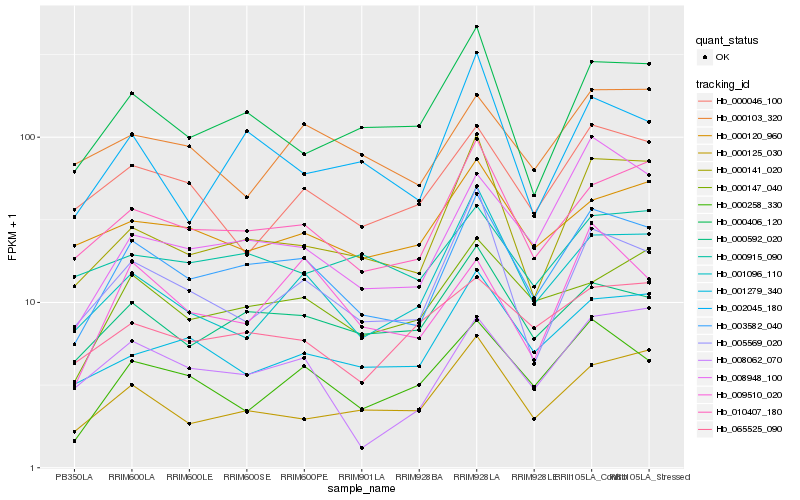
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003582_040 |
0.0 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0010s11160g [Populus trichocarpa] |
| 2 |
Hb_010407_180 |
0.1030498623 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 3 |
Hb_000592_020 |
0.1113903921 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 4 |
Hb_000141_020 |
0.124034694 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 6 [Pyrus x bretschneideri] |
| 5 |
Hb_000147_040 |
0.1241418522 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 6 |
Hb_000258_330 |
0.1251786878 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 7 |
Hb_005569_020 |
0.1268710521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000406_120 |
0.1327787486 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_008062_070 |
0.1336202757 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |
| 10 |
Hb_001279_340 |
0.1339922679 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 11 |
Hb_000125_030 |
0.1350446559 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 12 |
Hb_001096_110 |
0.136551358 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 13 |
Hb_002045_180 |
0.1391879077 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 14 |
Hb_008948_100 |
0.1396419489 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 15 |
Hb_000120_960 |
0.1410626861 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000046_100 |
0.141649542 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_065525_090 |
0.1459163058 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 18 |
Hb_009510_020 |
0.1480670532 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 19 |
Hb_000103_320 |
0.1508059713 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 20 |
Hb_000915_090 |
0.1513409277 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b [Jatropha curcas] |