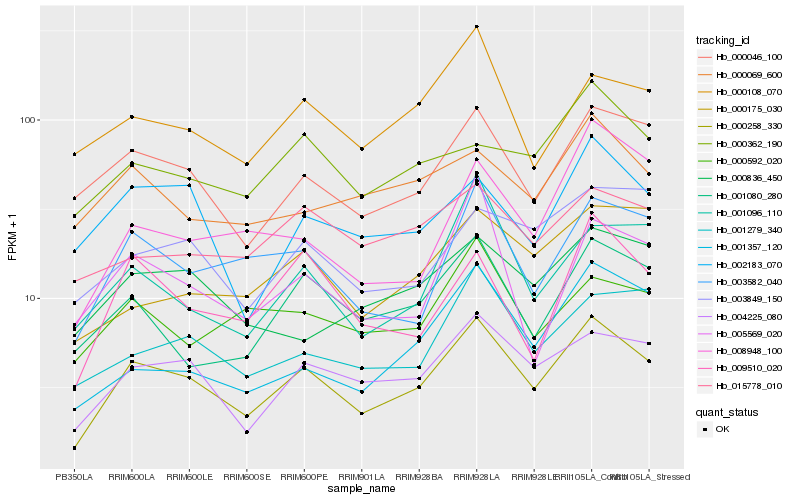
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_330 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_004225_080 |
0.1075275822 |
- |
- |
- |
| 3 |
Hb_000046_100 |
0.1219005607 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_003582_040 |
0.1251786878 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0010s11160g [Populus trichocarpa] |
| 5 |
Hb_001080_280 |
0.1355724146 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 6 |
Hb_009510_020 |
0.139872118 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 7 |
Hb_001279_340 |
0.1427733578 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 8 |
Hb_000108_070 |
0.1445814945 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 9 |
Hb_002183_070 |
0.1457521549 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001096_110 |
0.1463468172 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 11 |
Hb_001357_120 |
0.1492499986 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 12 |
Hb_000175_030 |
0.1520472662 |
- |
- |
Signal recognition particle, SRP54 subunit protein [Theobroma cacao] |
| 13 |
Hb_000592_020 |
0.1521926098 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 14 |
Hb_003849_150 |
0.1527158195 |
- |
- |
PREDICTED: uncharacterized protein LOC105644698 [Jatropha curcas] |
| 15 |
Hb_000836_450 |
0.1529956682 |
- |
- |
PREDICTED: 3-methyl-2-oxobutanoate hydroxymethyltransferase 1, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000362_190 |
0.1545218497 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 17 |
Hb_000069_600 |
0.1549873055 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 18 |
Hb_005569_020 |
0.1554638693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_015778_010 |
0.1556757213 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 1 [Jatropha curcas] |
| 20 |
Hb_008948_100 |
0.1563308651 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |