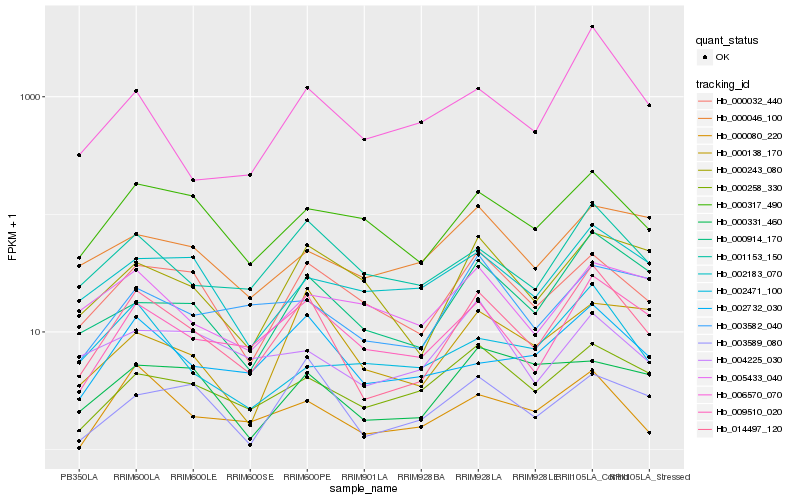
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014497_120 |
0.0 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 2 |
Hb_009510_020 |
0.1481480377 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 3 |
Hb_001153_150 |
0.1687436936 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 4 |
Hb_000914_170 |
0.1750740963 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 5 |
Hb_000032_440 |
0.1754884159 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_002732_030 |
0.177539876 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000258_330 |
0.1847817026 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_000317_490 |
0.1906308788 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 9 |
Hb_000080_220 |
0.1931317765 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000243_080 |
0.1934691703 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 11 |
Hb_006570_070 |
0.1982142511 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 12 |
Hb_003589_080 |
0.2084418857 |
- |
- |
PREDICTED: uncharacterized protein LOC105107863 [Populus euphratica] |
| 13 |
Hb_002183_070 |
0.2114661259 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_003582_040 |
0.212641241 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0010s11160g [Populus trichocarpa] |
| 15 |
Hb_000138_170 |
0.216457943 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 16 |
Hb_004225_030 |
0.2216905383 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 17 |
Hb_000331_460 |
0.2218493654 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 18 |
Hb_005433_040 |
0.2232620772 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 19 |
Hb_000046_100 |
0.2245688408 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_002471_100 |
0.2275248798 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |