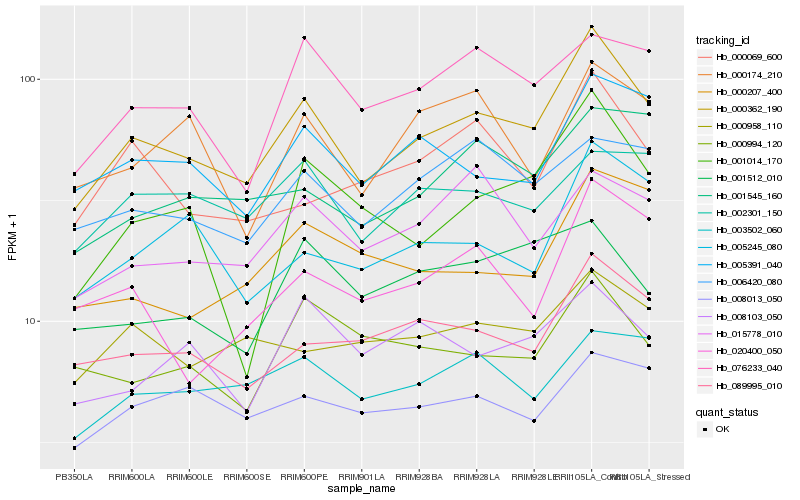
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_190 |
0.0 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 2 |
Hb_089995_010 |
0.1018131824 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like [Eucalyptus grandis] |
| 3 |
Hb_005391_040 |
0.1047679323 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA thioesterase 1 [Jatropha curcas] |
| 4 |
Hb_076233_040 |
0.1127039942 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 5 |
Hb_003502_060 |
0.1132036667 |
- |
- |
PREDICTED: alpha-1,3/1,6-mannosyltransferase ALG2 [Jatropha curcas] |
| 6 |
Hb_001545_160 |
0.1137408918 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC102615208 isoform X1 [Citrus sinensis] |
| 7 |
Hb_006420_080 |
0.1142943198 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_005245_080 |
0.1144963214 |
- |
- |
PREDICTED: uncharacterized protein LOC105643645 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000958_110 |
0.1166241125 |
- |
- |
PREDICTED: uncharacterized protein LOC105628719 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000069_600 |
0.117062397 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 11 |
Hb_015778_010 |
0.1180733229 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 1 [Jatropha curcas] |
| 12 |
Hb_002301_150 |
0.1189776634 |
- |
- |
Drought-responsive family protein [Theobroma cacao] |
| 13 |
Hb_008013_050 |
0.1208104425 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420 [Jatropha curcas] |
| 14 |
Hb_020400_050 |
0.1209174029 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000207_400 |
0.1209802142 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase PTP1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000994_120 |
0.1214195597 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_008103_050 |
0.1216468879 |
- |
- |
hypothetical protein JCGZ_07422 [Jatropha curcas] |
| 18 |
Hb_000174_210 |
0.1221615304 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 19 |
Hb_001014_170 |
0.1226432243 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001512_010 |
0.1232850139 |
- |
- |
PREDICTED: uncharacterized protein LOC105633759 [Jatropha curcas] |