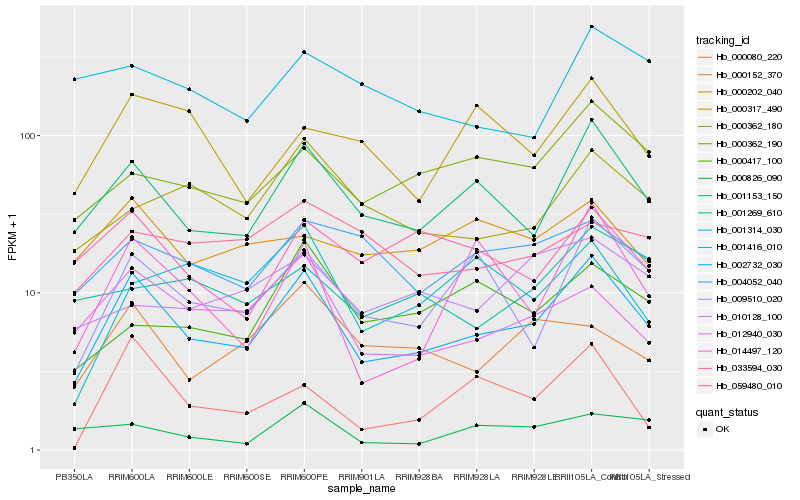
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002732_030 |
0.0 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001153_150 |
0.1169188135 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 3 |
Hb_012940_030 |
0.1581934033 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 4 |
Hb_009510_020 |
0.1609945345 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 5 |
Hb_000362_190 |
0.1754982896 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 6 |
Hb_014497_120 |
0.177539876 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 7 |
Hb_000362_180 |
0.1850220986 |
- |
- |
Pdap1 [Gossypium arboreum] |
| 8 |
Hb_004052_040 |
0.187631585 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 9 |
Hb_000152_370 |
0.1884734588 |
- |
- |
PREDICTED: uncharacterized protein LOC105648579 isoform X1 [Jatropha curcas] |
| 10 |
Hb_033594_030 |
0.1897145662 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 11 |
Hb_000317_490 |
0.1946544001 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 12 |
Hb_000202_040 |
0.1947780616 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 13 |
Hb_001269_610 |
0.2015729409 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000417_100 |
0.2020610421 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 15 |
Hb_001314_030 |
0.2033564558 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 16 |
Hb_059480_010 |
0.2033923833 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 17 |
Hb_000826_090 |
0.2035906516 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |
| 18 |
Hb_001416_010 |
0.2038390617 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 19 |
Hb_010128_100 |
0.2045966862 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000080_220 |
0.2046931578 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |