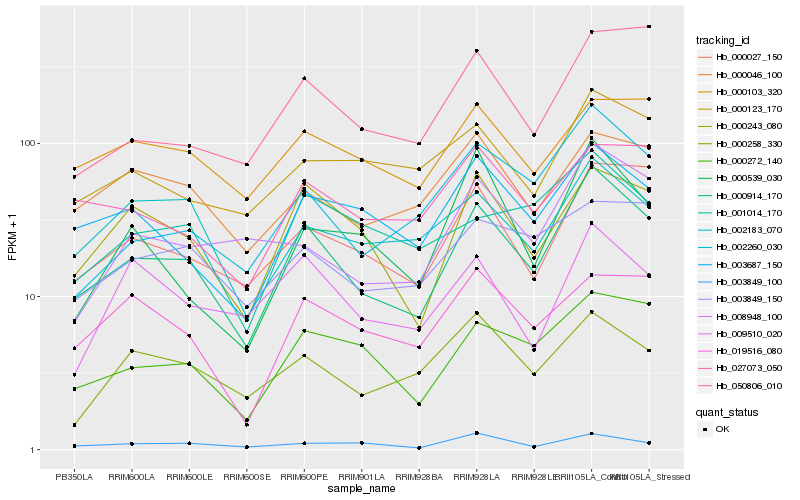
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000914_170 |
0.0 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 2 |
Hb_000243_080 |
0.1283890258 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 3 |
Hb_000272_140 |
0.1334818502 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 4 |
Hb_000027_150 |
0.1355017267 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 5 |
Hb_003687_150 |
0.1363314888 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002260_030 |
0.1385982542 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 7 |
Hb_000046_100 |
0.1555350245 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000123_170 |
0.1564338553 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 9 |
Hb_050806_010 |
0.1572379729 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 10 |
Hb_003849_100 |
0.1574319194 |
- |
- |
PREDICTED: putative transcription elongation factor SPT5 homolog 1 [Populus euphratica] |
| 11 |
Hb_000258_330 |
0.1575547986 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 12 |
Hb_000103_320 |
0.158044996 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 13 |
Hb_008948_100 |
0.158153388 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 14 |
Hb_002183_070 |
0.1593500199 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001014_170 |
0.1596822319 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_027073_050 |
0.160220439 |
- |
- |
PREDICTED: uncharacterized protein LOC105635813 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003849_150 |
0.1604874401 |
- |
- |
PREDICTED: uncharacterized protein LOC105644698 [Jatropha curcas] |
| 18 |
Hb_019516_080 |
0.1611609419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000539_030 |
0.1642215721 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
diphosphomevelonate decarboxylase [Hevea brasiliensis] |
| 20 |
Hb_009510_020 |
0.1642404293 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |