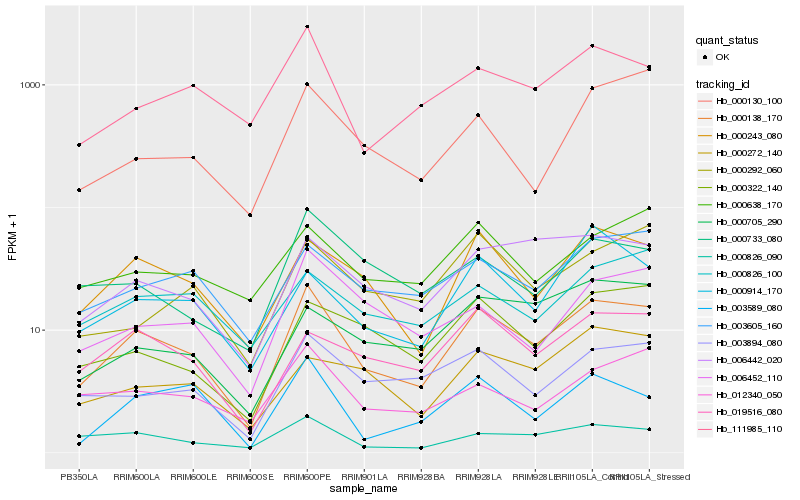
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000138_170 |
0.0 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 2 |
Hb_000243_080 |
0.1312456761 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 3 |
Hb_000826_090 |
0.1471138693 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |
| 4 |
Hb_006442_020 |
0.1497434395 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 5 |
Hb_003894_080 |
0.1512361888 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Vitis vinifera] |
| 6 |
Hb_111985_110 |
0.1562170133 |
- |
- |
thioredoxin H-type 3 [Hevea brasiliensis] |
| 7 |
Hb_019516_080 |
0.157050374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003605_160 |
0.1579560443 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 9 |
Hb_000322_140 |
0.160787976 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 10 |
Hb_012340_050 |
0.1622954298 |
- |
- |
PREDICTED: actin-related protein 5 [Jatropha curcas] |
| 11 |
Hb_006452_110 |
0.1636457245 |
- |
- |
Ubiquinol-cytochrome c reductase complex 7.8 kDa protein, putative [Ricinus communis] |
| 12 |
Hb_003589_080 |
0.1661130828 |
- |
- |
PREDICTED: uncharacterized protein LOC105107863 [Populus euphratica] |
| 13 |
Hb_000914_170 |
0.1701965318 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 14 |
Hb_000733_080 |
0.1702406138 |
- |
- |
PREDICTED: uncharacterized protein LOC105646960 [Jatropha curcas] |
| 15 |
Hb_000130_100 |
0.1710944218 |
- |
- |
hypothetical protein CICLE_v10022662mg [Citrus clementina] |
| 16 |
Hb_000638_170 |
0.172168319 |
- |
- |
PREDICTED: ras-related protein RABC1 [Jatropha curcas] |
| 17 |
Hb_000292_060 |
0.1733557511 |
- |
- |
transporter, putative [Ricinus communis] |
| 18 |
Hb_000272_140 |
0.1741448574 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 19 |
Hb_000826_100 |
0.1749993374 |
- |
- |
PREDICTED: biogenesis of lysosome-related organelles complex 1 subunit 1 [Jatropha curcas] |
| 20 |
Hb_000705_290 |
0.1761422803 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |