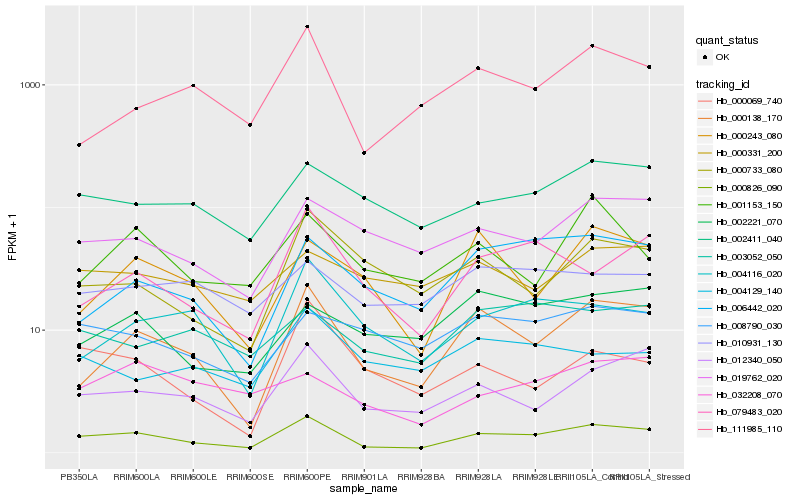
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000826_090 |
0.0 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |
| 2 |
Hb_000138_170 |
0.1471138693 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 3 |
Hb_012340_050 |
0.1499574047 |
- |
- |
PREDICTED: actin-related protein 5 [Jatropha curcas] |
| 4 |
Hb_002411_040 |
0.1593312032 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 6 [Jatropha curcas] |
| 5 |
Hb_019762_020 |
0.1639480661 |
- |
- |
PREDICTED: selenoprotein K-like [Nicotiana tomentosiformis] |
| 6 |
Hb_111985_110 |
0.1707685665 |
- |
- |
thioredoxin H-type 3 [Hevea brasiliensis] |
| 7 |
Hb_079483_020 |
0.17400807 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_006442_020 |
0.1762608267 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 9 |
Hb_008790_030 |
0.1794690928 |
- |
- |
PREDICTED: probable magnesium transporter NIPA9 [Jatropha curcas] |
| 10 |
Hb_000733_080 |
0.1811491167 |
- |
- |
PREDICTED: uncharacterized protein LOC105646960 [Jatropha curcas] |
| 11 |
Hb_003052_050 |
0.1811527973 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 12 |
Hb_004116_020 |
0.182079967 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 13 |
Hb_010931_130 |
0.1867043225 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 14 |
Hb_002221_070 |
0.1876651875 |
- |
- |
beta-glucanase, putative [Ricinus communis] |
| 15 |
Hb_000069_740 |
0.188303207 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 16 |
Hb_000331_200 |
0.1892965551 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 17 |
Hb_001153_150 |
0.1898981727 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 18 |
Hb_032208_070 |
0.190681829 |
- |
- |
PREDICTED: probable polyamine transporter At3g13620 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000243_080 |
0.1917947153 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 20 |
Hb_004129_140 |
0.1918810418 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |