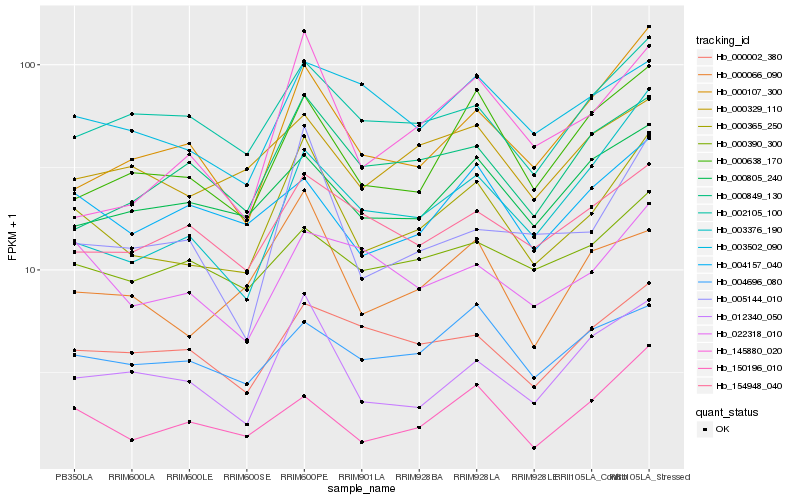
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000066_090 |
0.1141076315 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 3 |
Hb_012340_050 |
0.1180205238 |
- |
- |
PREDICTED: actin-related protein 5 [Jatropha curcas] |
| 4 |
Hb_000638_170 |
0.1237817894 |
- |
- |
PREDICTED: ras-related protein RABC1 [Jatropha curcas] |
| 5 |
Hb_005144_010 |
0.1241392868 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 6 |
Hb_002105_100 |
0.125013891 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_000107_300 |
0.1271583317 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 [Jatropha curcas] |
| 8 |
Hb_000329_110 |
0.1305707767 |
- |
- |
PREDICTED: uncharacterized protein LOC105643142 [Jatropha curcas] |
| 9 |
Hb_150196_010 |
0.1319723598 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 10 |
Hb_145880_020 |
0.1341469377 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 11 |
Hb_000002_380 |
0.1346111449 |
- |
- |
PREDICTED: bet1-like SNARE 1-2 [Jatropha curcas] |
| 12 |
Hb_003376_190 |
0.1352577501 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 13 |
Hb_022318_010 |
0.1356434767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000805_240 |
0.1359576609 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 15 |
Hb_004696_080 |
0.1361446328 |
- |
- |
PREDICTED: uncharacterized protein At1g01500-like [Populus euphratica] |
| 16 |
Hb_154948_040 |
0.136183534 |
- |
- |
hypothetical protein B456_013G043800 [Gossypium raimondii] |
| 17 |
Hb_004157_040 |
0.136472136 |
- |
- |
Rab2 [Hevea brasiliensis] |
| 18 |
Hb_000849_130 |
0.1369032009 |
- |
- |
PREDICTED: adrenodoxin-like protein, mitochondrial [Nelumbo nucifera] |
| 19 |
Hb_000390_300 |
0.1370864643 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase MCC1 [Jatropha curcas] |
| 20 |
Hb_003502_090 |
0.1384136119 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |