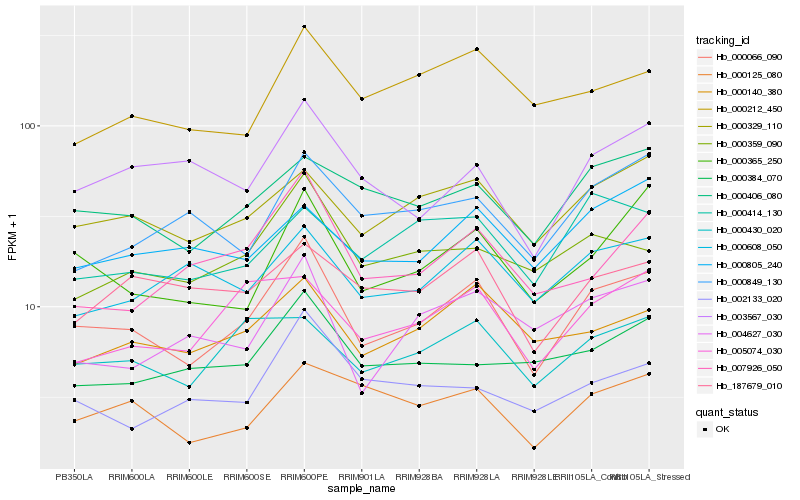
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000066_090 |
0.0 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 2 |
Hb_000365_250 |
0.1141076315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000329_110 |
0.1188682312 |
- |
- |
PREDICTED: uncharacterized protein LOC105643142 [Jatropha curcas] |
| 4 |
Hb_000406_080 |
0.1253924248 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 6B [Jatropha curcas] |
| 5 |
Hb_000140_380 |
0.1286680859 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 6 |
Hb_007926_050 |
0.1318797481 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 7 |
Hb_000359_090 |
0.1324552032 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 8 |
Hb_000125_080 |
0.1327910771 |
transcription factor |
TF Family: mTERF |
- |
| 9 |
Hb_000430_020 |
0.1336918844 |
- |
- |
- |
| 10 |
Hb_004627_030 |
0.1340608858 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 11 |
Hb_005074_030 |
0.1351827082 |
- |
- |
- |
| 12 |
Hb_003567_030 |
0.1353545237 |
- |
- |
PREDICTED: nifU-like protein 4, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000212_450 |
0.1392354776 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 14 |
Hb_000384_070 |
0.1397749287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000608_050 |
0.1402080337 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |
| 16 |
Hb_002133_020 |
0.1422332549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000849_130 |
0.1432048604 |
- |
- |
PREDICTED: adrenodoxin-like protein, mitochondrial [Nelumbo nucifera] |
| 18 |
Hb_000805_240 |
0.1438701813 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 19 |
Hb_187679_010 |
0.144036126 |
- |
- |
- |
| 20 |
Hb_000414_130 |
0.1452727323 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |