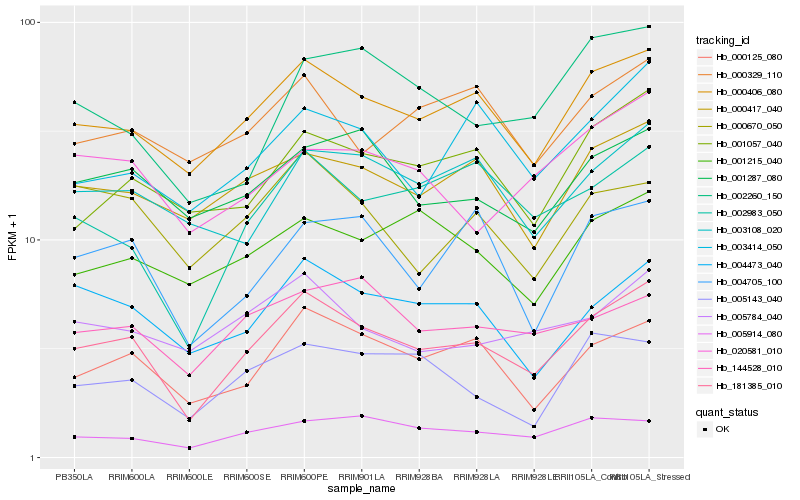
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_181385_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650319 [Jatropha curcas] |
| 2 |
Hb_000406_080 |
0.0851787263 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 6B [Jatropha curcas] |
| 3 |
Hb_000125_080 |
0.102360759 |
transcription factor |
TF Family: mTERF |
- |
| 4 |
Hb_004473_040 |
0.1155198347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002983_050 |
0.1171039894 |
- |
- |
PREDICTED: acetylornithine aminotransferase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000417_040 |
0.1189806214 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 7 |
Hb_001057_040 |
0.1202384822 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 8 |
Hb_005914_080 |
0.1234535753 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004705_100 |
0.1247675234 |
- |
- |
PREDICTED: inactive leucine-rich repeat receptor-like protein kinase CORYNE [Jatropha curcas] |
| 10 |
Hb_001287_080 |
0.1247981007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003414_050 |
0.1248219358 |
- |
- |
PREDICTED: uncharacterized protein LOC105640180 isoform X3 [Jatropha curcas] |
| 12 |
Hb_003108_020 |
0.1264081226 |
- |
- |
PREDICTED: transmembrane protein 18 [Jatropha curcas] |
| 13 |
Hb_005784_040 |
0.1297020696 |
- |
- |
PREDICTED: uncharacterized protein LOC105645160 isoform X1 [Jatropha curcas] |
| 14 |
Hb_144528_010 |
0.1313182493 |
- |
- |
type II inositol 5-phosphatase, putative [Ricinus communis] |
| 15 |
Hb_005143_040 |
0.1328395479 |
- |
- |
hypothetical protein JCGZ_17315 [Jatropha curcas] |
| 16 |
Hb_002260_150 |
0.1329040929 |
- |
- |
PREDICTED: uncharacterized protein LOC105647187 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000329_110 |
0.1330442294 |
- |
- |
PREDICTED: uncharacterized protein LOC105643142 [Jatropha curcas] |
| 18 |
Hb_020581_010 |
0.1330885471 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000670_050 |
0.1331297022 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 20 |
Hb_001215_040 |
0.1336218677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |