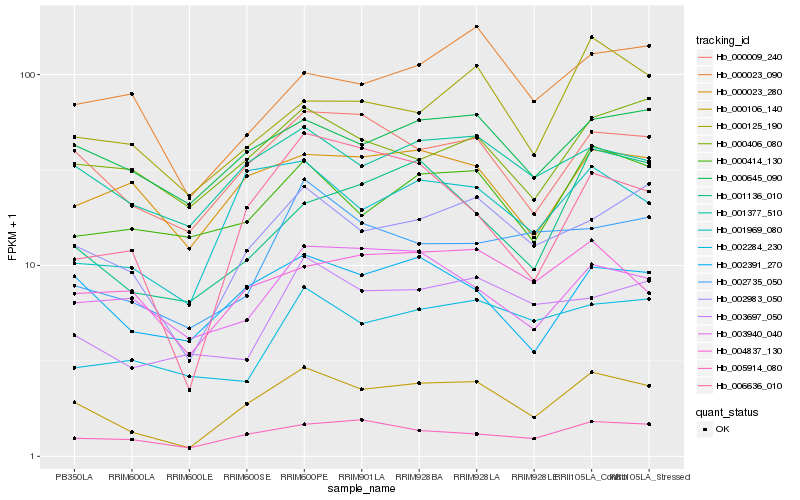
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000106_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_002983_050 |
0.1070812107 |
- |
- |
PREDICTED: acetylornithine aminotransferase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000009_240 |
0.113291431 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001969_080 |
0.1261761146 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |
| 5 |
Hb_001377_510 |
0.1295851728 |
- |
- |
ormdl, putative [Ricinus communis] |
| 6 |
Hb_002391_270 |
0.1307037204 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 7 |
Hb_005914_080 |
0.1308952363 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000645_090 |
0.1331411468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006636_010 |
0.1444123724 |
- |
- |
PREDICTED: uncharacterized protein LOC105637987 [Jatropha curcas] |
| 10 |
Hb_000414_130 |
0.1470513637 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 11 |
Hb_000023_280 |
0.1472393782 |
- |
- |
hypothetical protein POPTR_0006s28210g [Populus trichocarpa] |
| 12 |
Hb_003697_050 |
0.14811099 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 13 |
Hb_000406_080 |
0.1493854799 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 6B [Jatropha curcas] |
| 14 |
Hb_004837_130 |
0.1496938945 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 15 |
Hb_000125_190 |
0.1510855143 |
- |
- |
PREDICTED: F-box protein SKIP16 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002735_050 |
0.1531848601 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003940_040 |
0.1540404031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001136_010 |
0.1540970424 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 19 |
Hb_000023_090 |
0.1572223236 |
- |
- |
mak, putative [Ricinus communis] |
| 20 |
Hb_002284_230 |
0.1573750403 |
- |
- |
PREDICTED: uncharacterized protein At5g43822 [Jatropha curcas] |