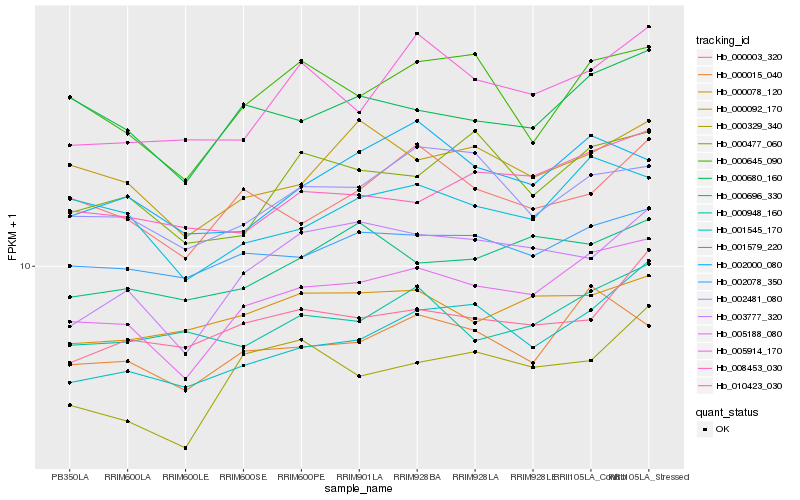
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005188_080 |
0.0 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 2 |
Hb_000015_040 |
0.0760229918 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000680_160 |
0.0760551123 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 4 |
Hb_002078_350 |
0.076693854 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 5 |
Hb_001545_170 |
0.077309063 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001579_220 |
0.0796692557 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |
| 7 |
Hb_000645_090 |
0.08042555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003777_320 |
0.081623625 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 15 homolog [Jatropha curcas] |
| 9 |
Hb_008453_030 |
0.0826612445 |
- |
- |
Thioredoxin superfamily protein [Theobroma cacao] |
| 10 |
Hb_002481_080 |
0.0859854645 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000477_060 |
0.087310455 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 12 |
Hb_000003_320 |
0.0880178077 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 13 |
Hb_000092_170 |
0.0886272181 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 14 |
Hb_000696_330 |
0.08883895 |
- |
- |
PREDICTED: uncharacterized protein LOC105647254 [Jatropha curcas] |
| 15 |
Hb_000078_120 |
0.0891581289 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 16 |
Hb_000329_340 |
0.08918154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005914_170 |
0.0894137784 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4E-2-like [Gossypium raimondii] |
| 18 |
Hb_000948_160 |
0.090099967 |
- |
- |
PREDICTED: 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_010423_030 |
0.0902489691 |
- |
- |
PREDICTED: uncharacterized protein LOC105645584 [Jatropha curcas] |
| 20 |
Hb_002000_080 |
0.0904069347 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |