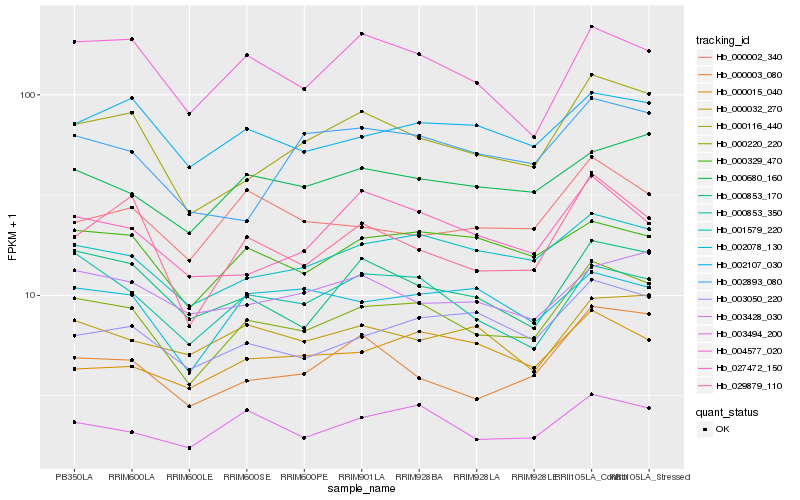
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 2 |
Hb_000116_440 |
0.0686656875 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 6-2 [Jatropha curcas] |
| 3 |
Hb_001579_220 |
0.072684135 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |
| 4 |
Hb_003494_200 |
0.0812092617 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 5 |
Hb_000680_160 |
0.0856611558 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 6 |
Hb_029879_110 |
0.0856773254 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000853_170 |
0.0875268656 |
- |
- |
PREDICTED: programmed cell death protein 2 isoform X3 [Populus euphratica] |
| 8 |
Hb_000329_470 |
0.0896210353 |
- |
- |
ccr4-not transcription complex, putative [Ricinus communis] |
| 9 |
Hb_003050_220 |
0.0901282985 |
- |
- |
hypothetical protein JCGZ_04601 [Jatropha curcas] |
| 10 |
Hb_002107_030 |
0.0916242478 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000003_080 |
0.0922915026 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000853_350 |
0.0927996711 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 13 |
Hb_004577_020 |
0.095651724 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 14 |
Hb_002078_130 |
0.0961402457 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 15 |
Hb_027472_150 |
0.0968473118 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 10 [Jatropha curcas] |
| 16 |
Hb_000002_340 |
0.0991695732 |
- |
- |
PREDICTED: peroxisomal bifunctional enzyme-like [Jatropha curcas] |
| 17 |
Hb_000032_270 |
0.1004694819 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 13 [Jatropha curcas] |
| 18 |
Hb_002893_080 |
0.1017634861 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |
| 19 |
Hb_003428_030 |
0.1021923004 |
- |
- |
tRNA, putative [Ricinus communis] |
| 20 |
Hb_000015_040 |
0.1022169784 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |