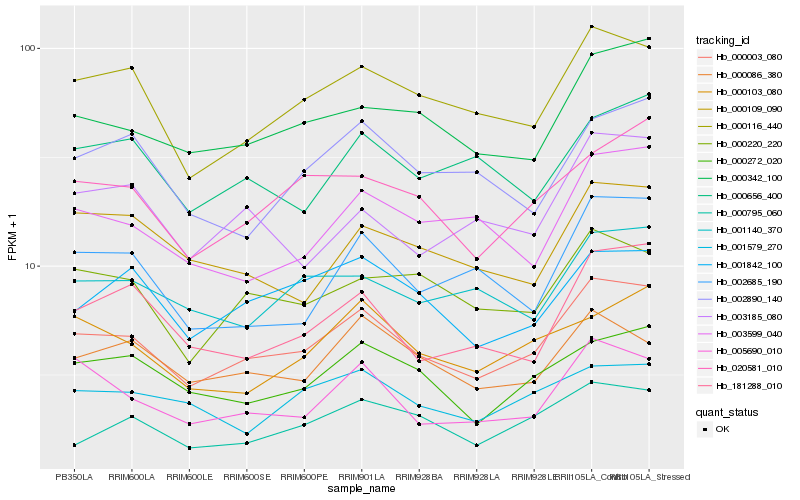
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_080 |
0.0 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000116_440 |
0.0745801721 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 6-2 [Jatropha curcas] |
| 3 |
Hb_000342_100 |
0.081111223 |
- |
- |
PREDICTED: PITH domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_020581_010 |
0.0872556033 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000272_020 |
0.089870017 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 6 |
Hb_000220_220 |
0.0922915026 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 7 |
Hb_001842_100 |
0.0934477251 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
hypothetical protein JCGZ_10905 [Jatropha curcas] |
| 8 |
Hb_181288_010 |
0.0947668965 |
- |
- |
rac-GTP binding protein, putative [Ricinus communis] |
| 9 |
Hb_005690_010 |
0.0948814788 |
- |
- |
PREDICTED: origin recognition complex subunit 2 [Populus euphratica] |
| 10 |
Hb_001140_370 |
0.0968014887 |
- |
- |
PREDICTED: uncharacterized protein LOC105648463 [Jatropha curcas] |
| 11 |
Hb_000086_380 |
0.0989438598 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000795_060 |
0.0991133615 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 13 |
Hb_003599_040 |
0.0993625379 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000103_080 |
0.1013673117 |
- |
- |
PREDICTED: ATP-dependent RNA helicase SUV3, mitochondrial [Jatropha curcas] |
| 15 |
Hb_002685_190 |
0.1025570654 |
transcription factor |
TF Family: SET |
PREDICTED: probable zinc metalloprotease EGY1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000656_400 |
0.1037715605 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 17 |
Hb_001579_270 |
0.1042606909 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 18 |
Hb_002890_140 |
0.104402818 |
- |
- |
PREDICTED: uncharacterized protein LOC100263805 [Vitis vinifera] |
| 19 |
Hb_000109_090 |
0.1060054744 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 20 |
Hb_003185_080 |
0.1064302927 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |