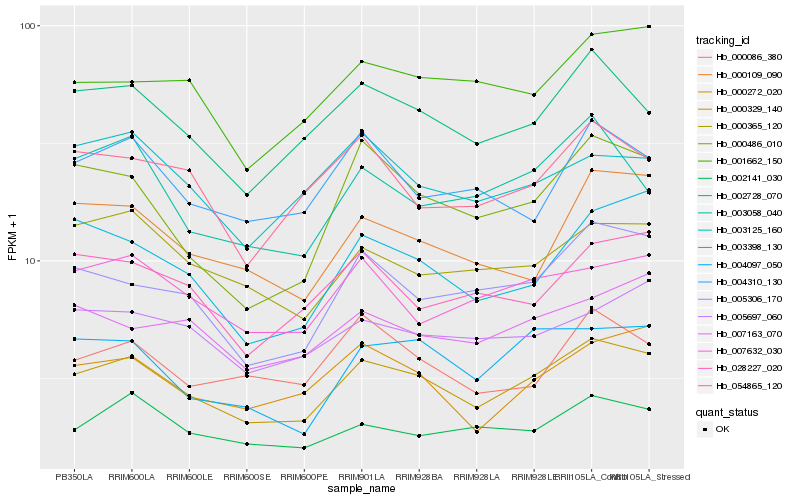
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_140 |
0.0 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 2 |
Hb_002728_070 |
0.0735020007 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 3 |
Hb_005306_170 |
0.0815809646 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 4 |
Hb_000272_020 |
0.0851416271 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 5 |
Hb_000365_120 |
0.0857439222 |
- |
- |
PREDICTED: bifunctional protein FolD 1, mitochondrial [Jatropha curcas] |
| 6 |
Hb_004097_050 |
0.0874861418 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |
| 7 |
Hb_007632_030 |
0.0879684203 |
- |
- |
PREDICTED: poly(A)-specific ribonuclease PARN isoform X1 [Jatropha curcas] |
| 8 |
Hb_002141_030 |
0.0883233498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000486_010 |
0.0893959659 |
- |
- |
PREDICTED: uncharacterized protein LOC105646661 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001662_150 |
0.0896156256 |
- |
- |
Scaffold attachment factor B1 [Medicago truncatula] |
| 11 |
Hb_000109_090 |
0.0911979632 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 12 |
Hb_003398_130 |
0.0932139456 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL30 [Gossypium raimondii] |
| 13 |
Hb_004310_130 |
0.0933074008 |
- |
- |
PREDICTED: uncharacterized protein LOC105643225 [Jatropha curcas] |
| 14 |
Hb_054865_120 |
0.0940487103 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 15 |
Hb_005697_060 |
0.094334581 |
transcription factor |
TF Family: HB |
PREDICTED: pathogenesis-related homeodomain protein isoform X1 [Jatropha curcas] |
| 16 |
Hb_028227_020 |
0.0945761071 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB isoform X1 [Jatropha curcas] |
| 17 |
Hb_003058_040 |
0.0946524465 |
- |
- |
rotamase, putative [Ricinus communis] |
| 18 |
Hb_003125_160 |
0.0947761446 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007163_070 |
0.0947934467 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000086_380 |
0.0949865471 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |