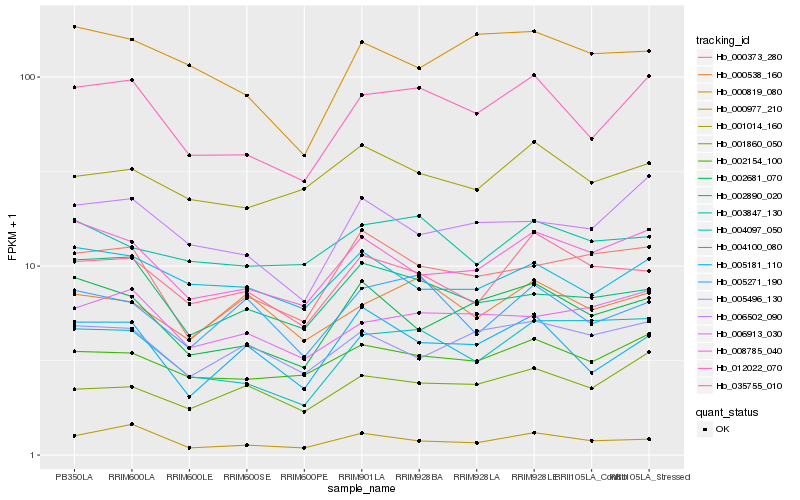
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012022_070 |
0.0 |
- |
- |
RecName: Full=Probable pyridoxal 5'-phosphate synthase subunit PDX1; Short=PLP synthase subunit PDX1; AltName: Full=Ethylene-inducible protein HEVER [Hevea brasiliensis] |
| 2 |
Hb_002154_100 |
0.089084044 |
- |
- |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 3 |
Hb_008785_040 |
0.095734028 |
- |
- |
PREDICTED: DNA repair protein UVH3 [Jatropha curcas] |
| 4 |
Hb_006502_090 |
0.0991701277 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 5 |
Hb_002681_070 |
0.1031372617 |
- |
- |
PREDICTED: uncharacterized protein LOC105633833 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004097_050 |
0.103882772 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |
| 7 |
Hb_006913_030 |
0.1053980706 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 8 |
Hb_005181_110 |
0.1063401437 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 9 |
Hb_035755_010 |
0.1070295352 |
- |
- |
unknown [Populus trichocarpa] |
| 10 |
Hb_002890_020 |
0.1097748898 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004100_080 |
0.1107060182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001860_050 |
0.112811882 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 13 |
Hb_000977_210 |
0.1144187079 |
- |
- |
chromatin assembly factor 1, subunit A, putative [Ricinus communis] |
| 14 |
Hb_005271_190 |
0.1150842765 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_000819_080 |
0.115311454 |
- |
- |
Plastid-lipid-associated protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001014_160 |
0.1157068111 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 17 |
Hb_000373_280 |
0.1161950199 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005496_130 |
0.1163933495 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000538_160 |
0.1164493667 |
- |
- |
PREDICTED: RNA-binding protein 5 isoform X3 [Jatropha curcas] |
| 20 |
Hb_003847_130 |
0.1178245404 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |