| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
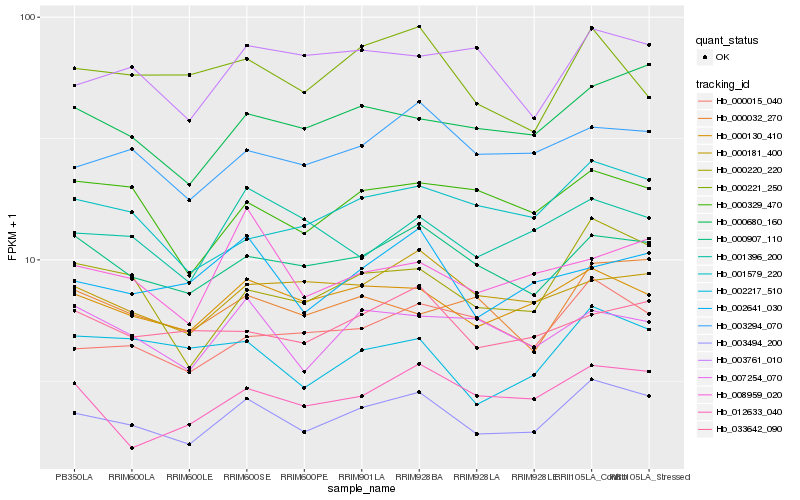
Hb_003494_200 |
0.0 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 2 |
Hb_000130_410 |
0.0753421804 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 3 |
Hb_000907_110 |
0.0796751407 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 4 |
Hb_000220_220 |
0.0812092617 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 5 |
Hb_000680_160 |
0.0863956385 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 6 |
Hb_003294_070 |
0.088889829 |
- |
- |
hypothetical protein RCOM_1714550 [Ricinus communis] |
| 7 |
Hb_000015_040 |
0.0892171991 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001579_220 |
0.093115282 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |
| 9 |
Hb_000181_400 |
0.0951407354 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002641_030 |
0.0962685041 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 11 |
Hb_007254_070 |
0.0964385485 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000329_470 |
0.0977710355 |
- |
- |
ccr4-not transcription complex, putative [Ricinus communis] |
| 13 |
Hb_002217_510 |
0.097894794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_033642_090 |
0.0980041968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012633_040 |
0.0983925454 |
- |
- |
- |
| 16 |
Hb_001396_200 |
0.0991018747 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000032_270 |
0.0995963285 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 13 [Jatropha curcas] |
| 18 |
Hb_003761_010 |
0.1012166251 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000221_250 |
0.102228587 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |
| 20 |
Hb_008959_020 |
0.1023478558 |
- |
- |
PREDICTED: nitric oxide synthase-interacting protein [Pyrus x bretschneideri] |