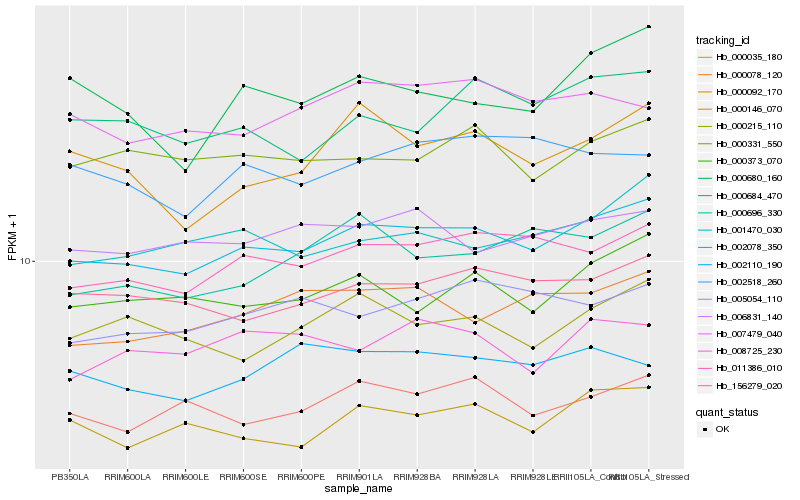
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_350 |
0.0 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 2 |
Hb_011386_010 |
0.0460271207 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 3 |
Hb_156279_020 |
0.052938592 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 4 |
Hb_000373_070 |
0.0572604638 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_006831_140 |
0.0585567569 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_001470_030 |
0.0586379053 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000035_180 |
0.0590865553 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 [Jatropha curcas] |
| 8 |
Hb_000331_550 |
0.0599122239 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 9 |
Hb_000078_120 |
0.0599726559 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 10 |
Hb_000146_070 |
0.0602030081 |
- |
- |
PREDICTED: RNA pseudouridine synthase 4, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000215_110 |
0.0614849026 |
- |
- |
PREDICTED: uncharacterized protein C3F10.06c isoform X1 [Jatropha curcas] |
| 12 |
Hb_005054_110 |
0.0634302272 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 13 |
Hb_007479_040 |
0.0659430142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000092_170 |
0.0665975653 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 15 |
Hb_000684_470 |
0.0666386356 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 16 |
Hb_000696_330 |
0.0674870704 |
- |
- |
PREDICTED: uncharacterized protein LOC105647254 [Jatropha curcas] |
| 17 |
Hb_008725_230 |
0.0676655664 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 18 |
Hb_000680_160 |
0.0680032048 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 19 |
Hb_002110_190 |
0.0689736363 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27460 [Jatropha curcas] |
| 20 |
Hb_002518_260 |
0.0694327049 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |