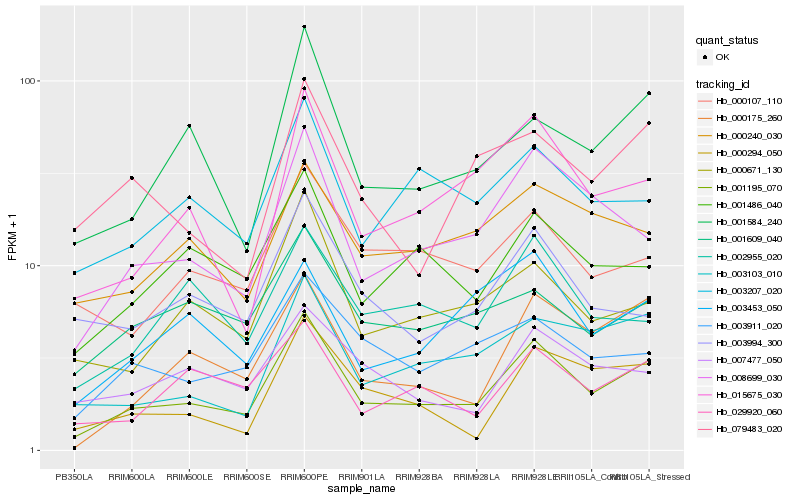
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_070 |
0.0 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 2 |
Hb_008699_030 |
0.1316922941 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 3 |
Hb_015675_030 |
0.1383285924 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000175_260 |
0.1408470483 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 5 |
Hb_001584_240 |
0.1418258628 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 6 |
Hb_003103_010 |
0.1425373951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000671_130 |
0.1473901337 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_029920_060 |
0.1502451371 |
- |
- |
PREDICTED: UPF0554 protein-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000294_050 |
0.1513415835 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 10 |
Hb_001486_040 |
0.152982716 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003911_020 |
0.1541643899 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 12 |
Hb_003207_020 |
0.1562326212 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 13 |
Hb_000240_030 |
0.1587711232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000107_110 |
0.1601046292 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_002955_020 |
0.1625706165 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_007477_050 |
0.162910163 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001609_040 |
0.1630015735 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |
| 18 |
Hb_079483_020 |
0.1641912726 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_003453_050 |
0.1654375872 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 20 |
Hb_003994_300 |
0.1674507018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |