| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
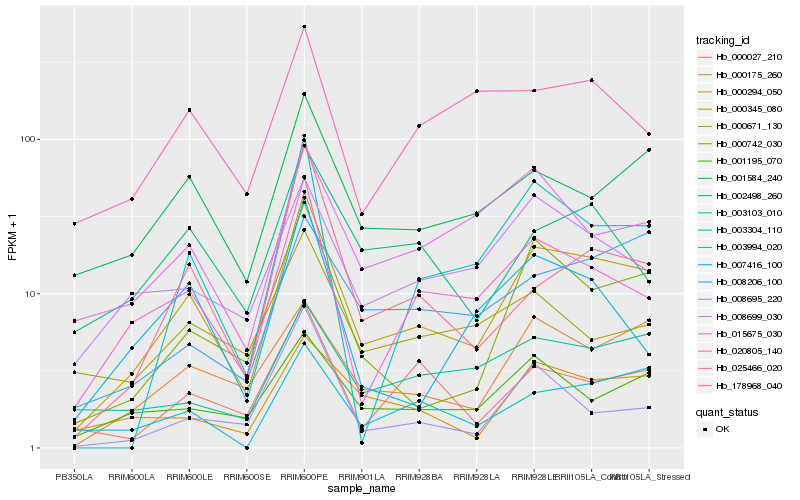
Hb_000345_080 |
0.0 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 2 |
Hb_000742_030 |
0.1532165126 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 3 |
Hb_020805_140 |
0.1548559488 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 4 |
Hb_001584_240 |
0.1662029079 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 5 |
Hb_000175_260 |
0.1675421325 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 6 |
Hb_000294_050 |
0.1752410213 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 7 |
Hb_003304_110 |
0.1784928918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001195_070 |
0.1795757307 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 9 |
Hb_008699_030 |
0.1813711273 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 10 |
Hb_003994_020 |
0.1819258975 |
- |
- |
- |
| 11 |
Hb_000027_210 |
0.1915241533 |
- |
- |
Os01g0689300, partial [Oryza sativa Japonica Group] |
| 12 |
Hb_178968_040 |
0.1918749877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_015675_030 |
0.1919920972 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003103_010 |
0.1928099839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008206_100 |
0.1935821123 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 16 |
Hb_002498_260 |
0.1952383765 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 17 |
Hb_007416_100 |
0.2014348847 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 18 |
Hb_025466_020 |
0.2020770168 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 19 |
Hb_008695_220 |
0.2056558033 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 20 |
Hb_000671_130 |
0.2085845842 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |