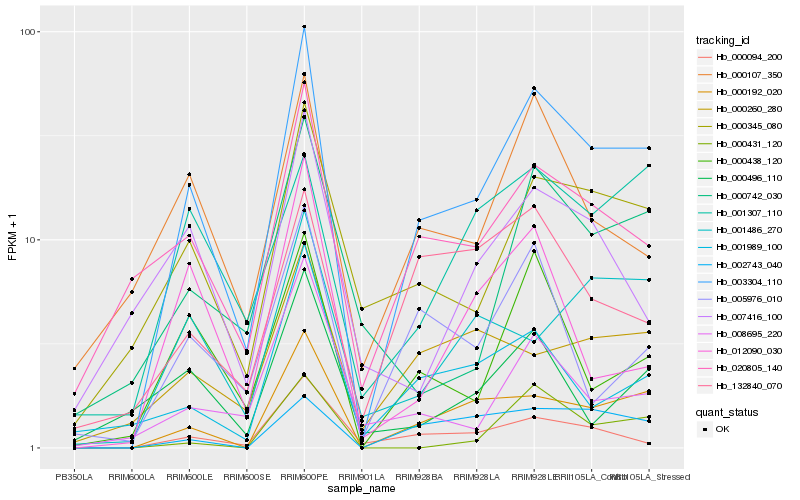
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003304_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000192_020 |
0.1519479661 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 3 |
Hb_000345_080 |
0.1784928918 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 4 |
Hb_020805_140 |
0.1850949716 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 5 |
Hb_000094_200 |
0.1971957821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000438_120 |
0.1991112283 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000107_350 |
0.2125296504 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 8 |
Hb_001989_100 |
0.2173902534 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 9 |
Hb_005976_010 |
0.2178412856 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000742_030 |
0.2198144436 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 11 |
Hb_002743_040 |
0.2215365043 |
- |
- |
PREDICTED: uncharacterized protein LOC105785507 isoform X2 [Gossypium raimondii] |
| 12 |
Hb_000260_280 |
0.221906621 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 13 |
Hb_008695_220 |
0.2219665156 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 14 |
Hb_001486_270 |
0.228254295 |
- |
- |
Ann10 [Manihot esculenta] |
| 15 |
Hb_000431_120 |
0.2297358872 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_001307_110 |
0.2345460796 |
- |
- |
PREDICTED: uncharacterized protein LOC105644507 [Jatropha curcas] |
| 17 |
Hb_000496_110 |
0.2346858135 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 18 |
Hb_132840_070 |
0.2370568107 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 19 |
Hb_012090_030 |
0.2378529268 |
- |
- |
PREDICTED: autophagy-related protein 8C isoform X2 [Phoenix dactylifera] |
| 20 |
Hb_007416_100 |
0.2389787758 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |