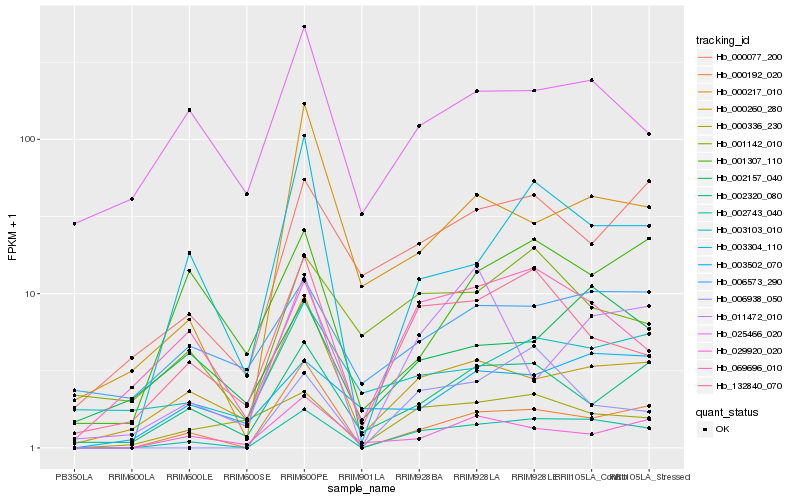
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002743_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105785507 isoform X2 [Gossypium raimondii] |
| 2 |
Hb_000192_020 |
0.1937077533 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 3 |
Hb_132840_070 |
0.2172438779 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 4 |
Hb_069696_010 |
0.2193760708 |
- |
- |
hypothetical protein POPTR_0003s21190g [Populus trichocarpa] |
| 5 |
Hb_003304_110 |
0.2215365043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002320_080 |
0.2293161662 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 7 |
Hb_006938_050 |
0.2349502027 |
- |
- |
PREDICTED: uncharacterized protein LOC105648204 [Jatropha curcas] |
| 8 |
Hb_000260_280 |
0.2437671051 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 9 |
Hb_000336_230 |
0.245160396 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa007609mg [Prunus persica] |
| 10 |
Hb_000077_200 |
0.2490195728 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 11 |
Hb_029920_020 |
0.2562788685 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 12 |
Hb_001142_010 |
0.2567518724 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |
| 13 |
Hb_025466_020 |
0.2597853383 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 14 |
Hb_002157_040 |
0.2599210893 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 15 |
Hb_003502_070 |
0.2613792938 |
- |
- |
hypothetical protein CISIN_1g0405401mg, partial [Citrus sinensis] |
| 16 |
Hb_006573_290 |
0.2635945119 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 17 |
Hb_001307_110 |
0.2659433686 |
- |
- |
PREDICTED: uncharacterized protein LOC105644507 [Jatropha curcas] |
| 18 |
Hb_000217_010 |
0.2674696638 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 19 |
Hb_011472_010 |
0.270096244 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 20 |
Hb_003103_010 |
0.2714264608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |