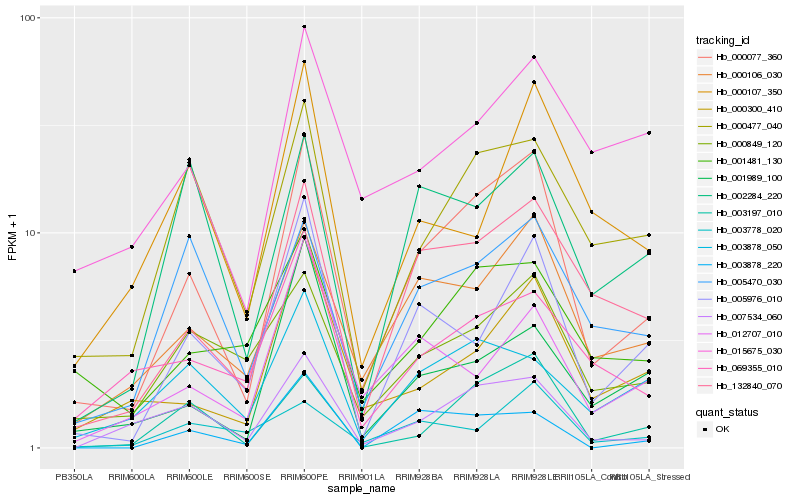
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_360 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_132840_070 |
0.1544296592 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 3 |
Hb_000106_030 |
0.1618137315 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_005976_010 |
0.1778044529 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007534_060 |
0.1797201732 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_003197_010 |
0.1797639426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000477_040 |
0.1886366627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000300_410 |
0.193112582 |
- |
- |
hypothetical protein POPTR_0013s05860g [Populus trichocarpa] |
| 9 |
Hb_000849_120 |
0.2016536844 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 10 |
Hb_001481_130 |
0.2051446332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005470_030 |
0.2080651085 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003878_050 |
0.2141063443 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 13 |
Hb_001989_100 |
0.2166551034 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 14 |
Hb_002284_220 |
0.2168658207 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 15 |
Hb_069355_010 |
0.217125521 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 16 |
Hb_012707_010 |
0.2180494916 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 17 |
Hb_015675_030 |
0.2196186672 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003878_220 |
0.2197213538 |
- |
- |
vacuolar protein sorting, putative [Ricinus communis] |
| 19 |
Hb_000107_350 |
0.219955692 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 20 |
Hb_003778_020 |
0.22185312 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |