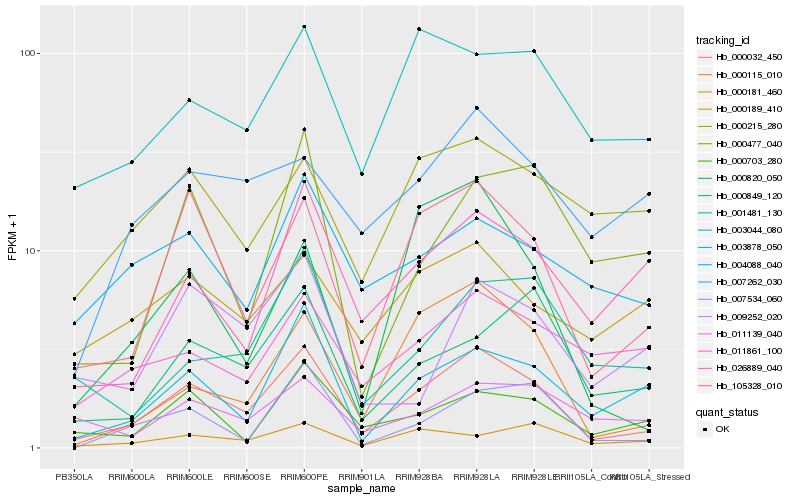
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_450 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 2 |
Hb_105328_010 |
0.1352009351 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 3 |
Hb_000115_010 |
0.1815819059 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_003878_050 |
0.1845680674 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 5 |
Hb_000703_280 |
0.1996698755 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 6 |
Hb_007534_060 |
0.1998080485 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_026889_040 |
0.2027171789 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 8 |
Hb_003044_080 |
0.2059636886 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 9 |
Hb_000849_120 |
0.2071387757 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 10 |
Hb_000820_050 |
0.208810789 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 11 |
Hb_001481_130 |
0.2107903319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004088_040 |
0.2132494736 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 13 |
Hb_011139_040 |
0.2142937668 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000181_460 |
0.2145969055 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 15 |
Hb_011861_100 |
0.2151992548 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_007262_030 |
0.2178285555 |
- |
- |
- |
| 17 |
Hb_009252_020 |
0.2206659533 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 18 |
Hb_000215_280 |
0.2208331627 |
- |
- |
Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor, putative [Ricinus communis] |
| 19 |
Hb_000477_040 |
0.2217674997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000189_410 |
0.2218263966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |