| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
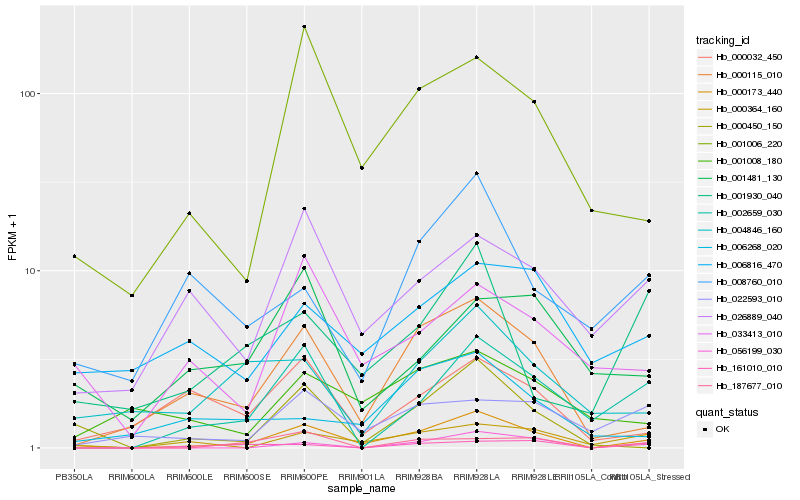
| 1 |
Hb_000173_440 |
0.0 |
- |
- |
YALI0B05280p [Yarrowia lipolytica] |
| 2 |
Hb_000115_010 |
0.2088840654 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000450_150 |
0.2333949758 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 4 |
Hb_001006_220 |
0.2357502355 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 5 |
Hb_000364_160 |
0.2519227024 |
- |
- |
PREDICTED: uncharacterized protein LOC105639754 [Jatropha curcas] |
| 6 |
Hb_002659_030 |
0.2564375811 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 7 |
Hb_056199_030 |
0.2635394155 |
- |
- |
PREDICTED: purine permease 3-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_033413_010 |
0.2748833221 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 9 |
Hb_004846_160 |
0.2757149765 |
- |
- |
- |
| 10 |
Hb_026889_040 |
0.2799469876 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 11 |
Hb_000032_450 |
0.2846120375 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 12 |
Hb_022593_010 |
0.2846815509 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_187677_010 |
0.2886784474 |
- |
- |
- |
| 14 |
Hb_001008_180 |
0.2888279425 |
- |
- |
- |
| 15 |
Hb_001930_040 |
0.2912924609 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_006816_470 |
0.2921177278 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008760_010 |
0.2925206995 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 18 |
Hb_006268_020 |
0.2967442917 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 19 |
Hb_001481_130 |
0.2990061056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_161010_010 |
0.3017014588 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |