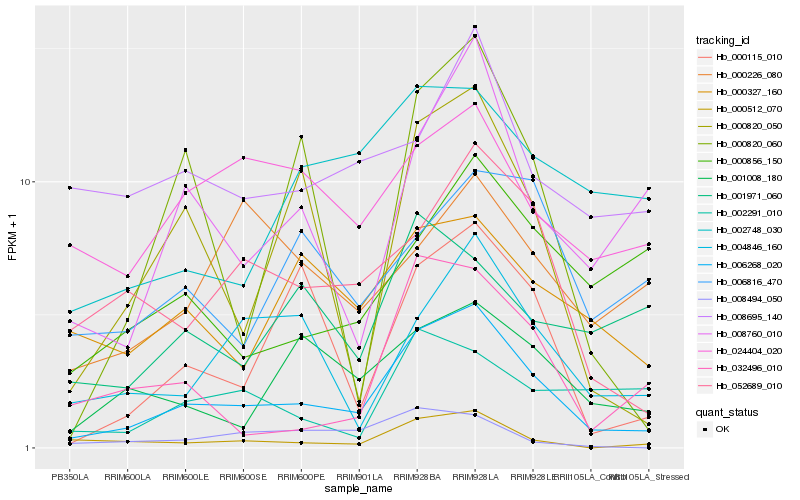
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006268_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 2 |
Hb_052689_010 |
0.1974784755 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 3 |
Hb_000512_070 |
0.2041349997 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71250-like [Jatropha curcas] |
| 4 |
Hb_000820_050 |
0.2070722676 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 5 |
Hb_008760_010 |
0.20898358 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 6 |
Hb_000115_010 |
0.2098657667 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_004846_160 |
0.2184186456 |
- |
- |
- |
| 8 |
Hb_001008_180 |
0.2185749137 |
- |
- |
- |
| 9 |
Hb_000327_160 |
0.2267406507 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_032496_010 |
0.2279452622 |
- |
- |
hypothetical protein POPTR_0012s10850g [Populus trichocarpa] |
| 11 |
Hb_002748_030 |
0.2342369036 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 12 |
Hb_000820_060 |
0.239481511 |
- |
- |
- |
| 13 |
Hb_008695_140 |
0.2422636871 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 14 |
Hb_024404_020 |
0.2433796646 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 15 |
Hb_000856_150 |
0.2438831232 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 16 |
Hb_006816_470 |
0.2473932237 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008494_050 |
0.2479090662 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 18 |
Hb_001971_060 |
0.252057016 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Populus euphratica] |
| 19 |
Hb_002291_010 |
0.2558785303 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_000226_080 |
0.2562299992 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |