| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
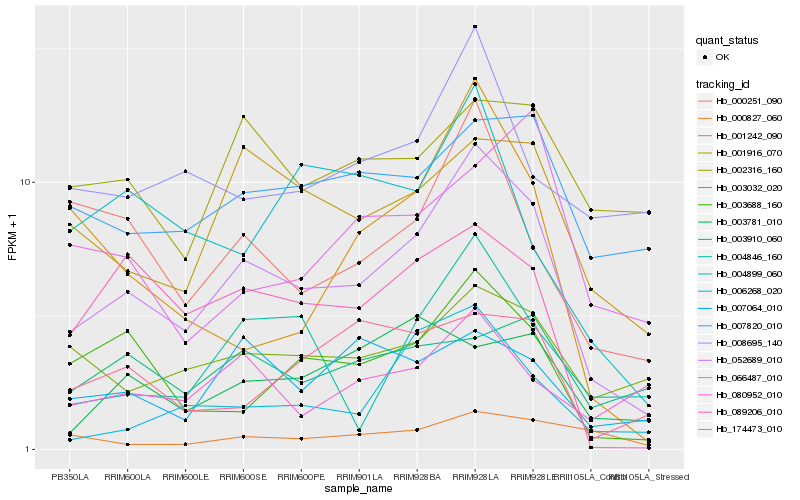
Hb_052689_010 |
0.0 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 2 |
Hb_001916_070 |
0.1783470356 |
- |
- |
PREDICTED: phytochrome C [Jatropha curcas] |
| 3 |
Hb_003688_160 |
0.1795882799 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 4 |
Hb_000251_090 |
0.1876644721 |
- |
- |
PREDICTED: two-pore potassium channel 3-like [Jatropha curcas] |
| 5 |
Hb_089206_010 |
0.1912570811 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 6 |
Hb_003032_020 |
0.1917885417 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55740, chloroplastic [Jatropha curcas] |
| 7 |
Hb_007820_010 |
0.1952625237 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: protein FLOWERING LOCUS D [Jatropha curcas] |
| 8 |
Hb_006268_020 |
0.1974784755 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 9 |
Hb_007064_010 |
0.2012370768 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 10 |
Hb_004846_160 |
0.2065472393 |
- |
- |
- |
| 11 |
Hb_008695_140 |
0.2074656266 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 12 |
Hb_000827_060 |
0.2105433562 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 13 |
Hb_174473_010 |
0.2113913758 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 14 |
Hb_080952_010 |
0.2122339574 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20770 [Jatropha curcas] |
| 15 |
Hb_004899_060 |
0.2126846564 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 16 |
Hb_001242_090 |
0.2136908734 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 17 |
Hb_002316_160 |
0.2142735474 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 18 |
Hb_003910_060 |
0.2157980826 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g37170 [Jatropha curcas] |
| 19 |
Hb_066487_010 |
0.2176746724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003781_010 |
0.219333366 |
- |
- |
PREDICTED: histone H2AX [Eucalyptus grandis] |