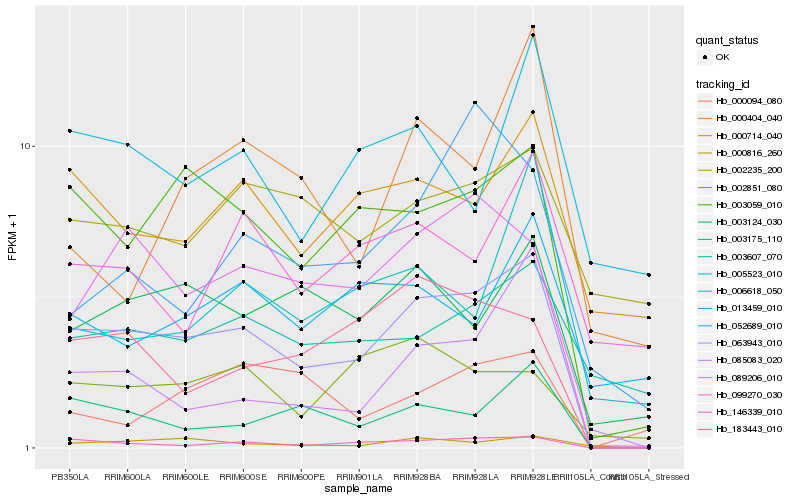
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063943_010 |
0.0 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 2 |
Hb_146339_010 |
0.132407085 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 3 |
Hb_003059_010 |
0.1405753375 |
- |
- |
PREDICTED: uncharacterized protein LOC104236538 [Nicotiana sylvestris] |
| 4 |
Hb_089206_010 |
0.1558827035 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 5 |
Hb_003175_110 |
0.1693453361 |
- |
- |
- |
| 6 |
Hb_183443_010 |
0.1821384495 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 7 |
Hb_003124_030 |
0.1861799556 |
- |
- |
At4g33625-like protein [Brassica napus] |
| 8 |
Hb_000816_260 |
0.1896304984 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_000714_040 |
0.2081882495 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002851_080 |
0.2096199554 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 11 |
Hb_085083_020 |
0.2112749259 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 12 |
Hb_003607_070 |
0.2198359055 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 13 |
Hb_005523_010 |
0.2262815321 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 14 |
Hb_052689_010 |
0.2280290208 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 15 |
Hb_099270_030 |
0.2291178987 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000094_080 |
0.2292061453 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 17 |
Hb_013459_010 |
0.2292521884 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 18 |
Hb_000404_040 |
0.230309889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006618_050 |
0.2306416484 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002235_200 |
0.23164064 |
- |
- |
pattern formation protein, putative [Ricinus communis] |