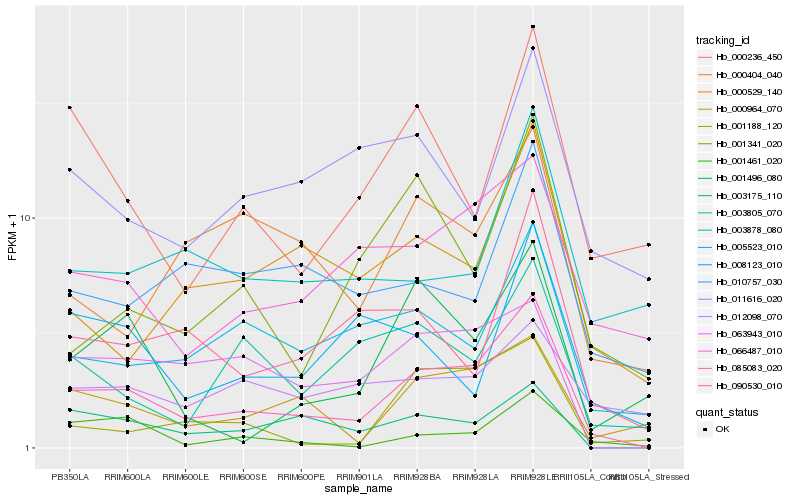
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_085083_020 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 2 |
Hb_003175_110 |
0.2029726244 |
- |
- |
- |
| 3 |
Hb_000964_070 |
0.2057804278 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 4 |
Hb_066487_010 |
0.2085495831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_063943_010 |
0.2112749259 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 6 |
Hb_005523_010 |
0.2119716232 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 7 |
Hb_090530_010 |
0.2222234191 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_001461_020 |
0.2233817441 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_000529_140 |
0.2266299217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003805_070 |
0.228079084 |
- |
- |
PREDICTED: sacsin [Jatropha curcas] |
| 11 |
Hb_000236_450 |
0.230009857 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 12 |
Hb_001188_120 |
0.2332488196 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 13 |
Hb_010757_030 |
0.2344873269 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001341_020 |
0.2348192948 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 15 |
Hb_001496_080 |
0.237222613 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 16 |
Hb_003878_080 |
0.2386381679 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |
| 17 |
Hb_000404_040 |
0.2391068096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011616_020 |
0.2412146948 |
- |
- |
hypothetical protein CICLE_v10004968mg [Citrus clementina] |
| 19 |
Hb_012098_070 |
0.2412905727 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 20 |
Hb_008123_010 |
0.241791977 |
- |
- |
- |