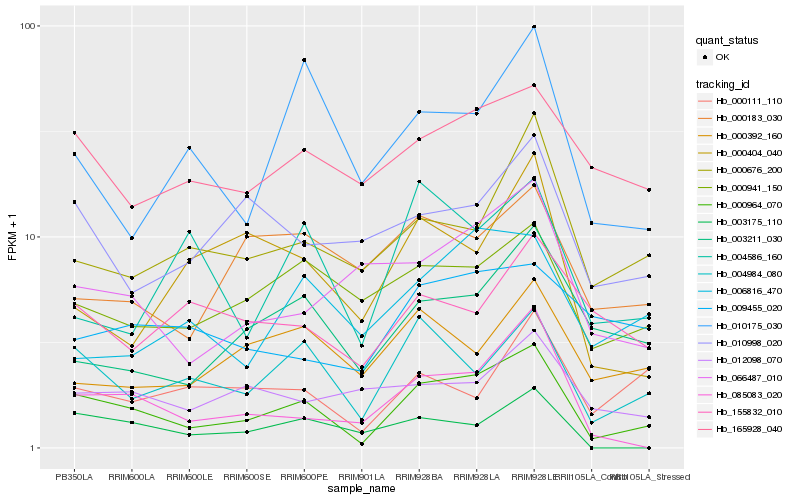
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000964_070 |
0.0 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 2 |
Hb_004984_080 |
0.184837239 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_085083_020 |
0.2057804278 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 4 |
Hb_000941_150 |
0.2194740518 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003211_030 |
0.2200550314 |
- |
- |
- |
| 6 |
Hb_000676_200 |
0.2215109532 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 7 |
Hb_066487_010 |
0.223473034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_165928_040 |
0.2283596863 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase component HRD3A [Jatropha curcas] |
| 9 |
Hb_000111_110 |
0.2319945747 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_010175_030 |
0.2329270262 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 11 |
Hb_010998_020 |
0.235475713 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 12 |
Hb_003175_110 |
0.2366054253 |
- |
- |
- |
| 13 |
Hb_006816_470 |
0.2410075701 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 14 |
Hb_012098_070 |
0.2413226718 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 15 |
Hb_000392_160 |
0.2413826538 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000404_040 |
0.2431869205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000183_030 |
0.2439104997 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0291141 [Populus euphratica] |
| 18 |
Hb_004586_160 |
0.2451589296 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_009455_020 |
0.2460037393 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 20 |
Hb_155832_010 |
0.2470580606 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |