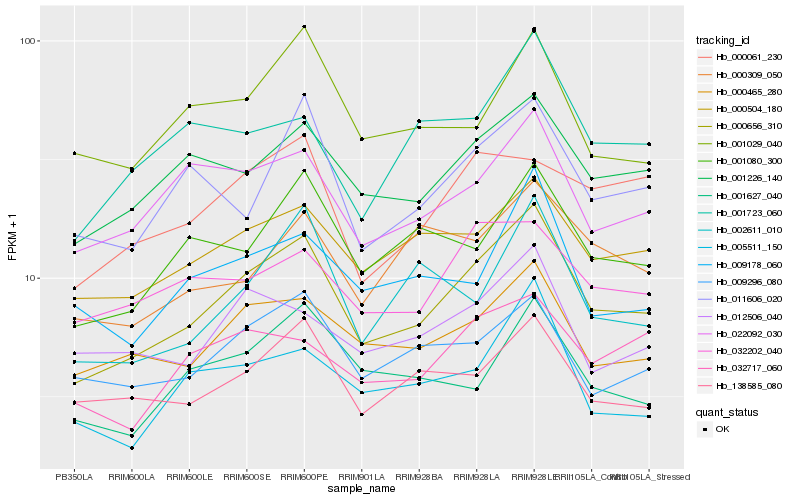
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_310 |
0.0 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 2 |
Hb_001226_140 |
0.1018781265 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 3 |
Hb_000465_280 |
0.1041360048 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 4 |
Hb_000504_180 |
0.1059459123 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 5 |
Hb_022092_030 |
0.1112661151 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001080_300 |
0.1124207833 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 7 |
Hb_138585_080 |
0.1160431442 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 8 |
Hb_011606_020 |
0.1166343432 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 9 |
Hb_009296_080 |
0.1178632081 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005511_150 |
0.1193211591 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic isoform X5 [Populus euphratica] |
| 11 |
Hb_001723_060 |
0.1225009368 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 12 |
Hb_009178_060 |
0.1227785039 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002611_010 |
0.1230297836 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_032717_060 |
0.1234530918 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 15 |
Hb_012506_040 |
0.1238455706 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 16 |
Hb_001627_040 |
0.1239467383 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001029_040 |
0.124895158 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 18 |
Hb_000061_230 |
0.1250211424 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 19 |
Hb_032202_040 |
0.1260563268 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 20 |
Hb_000309_050 |
0.1262030107 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |