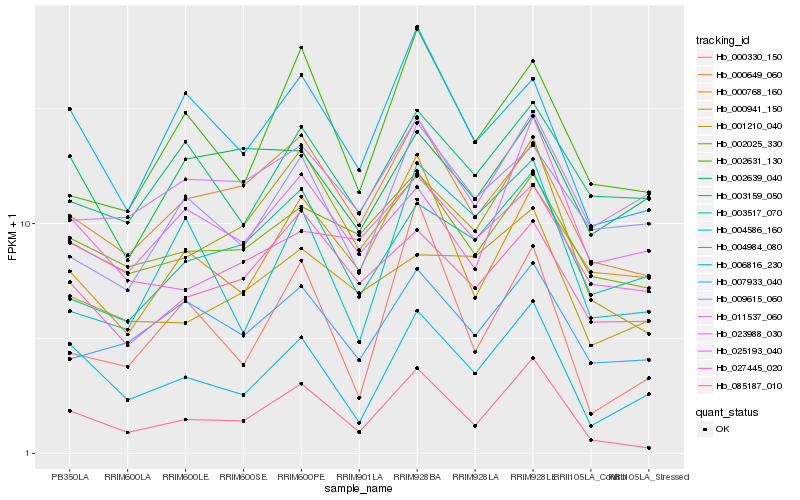
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004984_080 |
0.0 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_085187_010 |
0.1409970311 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_006816_230 |
0.1444806889 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 4 |
Hb_007933_040 |
0.166920687 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001210_040 |
0.1670510069 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 6 |
Hb_000649_060 |
0.1673615641 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 7 |
Hb_002639_040 |
0.1700926024 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 8 |
Hb_002025_330 |
0.1710828489 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 9 |
Hb_003159_050 |
0.1715208229 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_023988_030 |
0.1716127196 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 11 |
Hb_004586_160 |
0.1724068363 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_025193_040 |
0.1734826569 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 13 |
Hb_009615_060 |
0.1743459101 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 14 |
Hb_011537_060 |
0.1751199074 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 15 |
Hb_002631_130 |
0.1764205317 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 16 |
Hb_003517_070 |
0.1771797233 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 17 |
Hb_000768_160 |
0.1774366486 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 18 |
Hb_000330_150 |
0.1774404985 |
- |
- |
PREDICTED: dihydroflavonol-4-reductase-like [Jatropha curcas] |
| 19 |
Hb_000941_150 |
0.1778597092 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 20 |
Hb_027445_020 |
0.1780526096 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |