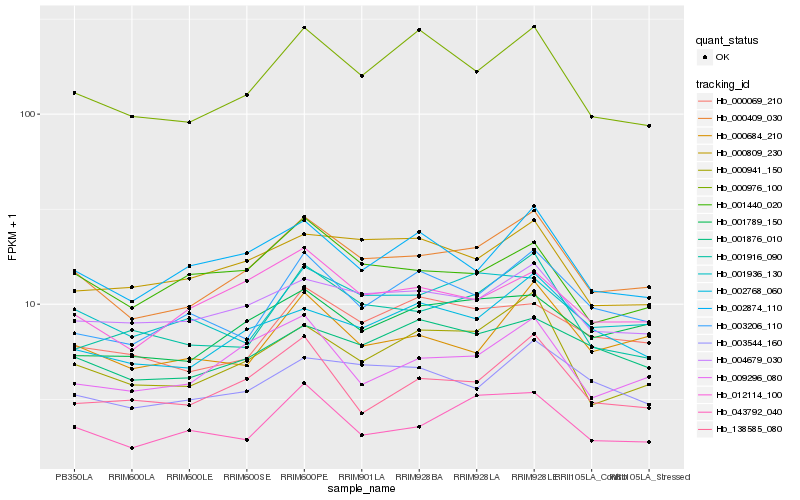
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000409_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642693 [Jatropha curcas] |
| 2 |
Hb_004679_030 |
0.0812306693 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 3 |
Hb_000941_150 |
0.0822229744 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002768_060 |
0.0839976289 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000976_100 |
0.0854500118 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 6 |
Hb_009296_080 |
0.0864418565 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_012114_100 |
0.0872259541 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 8 |
Hb_001789_150 |
0.0874066666 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003544_160 |
0.0877688835 |
- |
- |
- |
| 10 |
Hb_002874_110 |
0.089757936 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 11 |
Hb_001876_010 |
0.0906863442 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000684_210 |
0.0912284614 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 13 |
Hb_001916_090 |
0.091479728 |
- |
- |
beta-1,2-n-acetylglucosaminyltransferase II, putative [Ricinus communis] |
| 14 |
Hb_043792_040 |
0.0922548333 |
- |
- |
hypothetical protein, partial [Rosa rugosa] |
| 15 |
Hb_001936_130 |
0.0944381799 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 16 |
Hb_001440_020 |
0.0949829771 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 17 |
Hb_003206_110 |
0.0955627331 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 18 |
Hb_138585_080 |
0.0959104343 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 19 |
Hb_000069_210 |
0.0960677312 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 20 |
Hb_000809_230 |
0.0961755074 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |