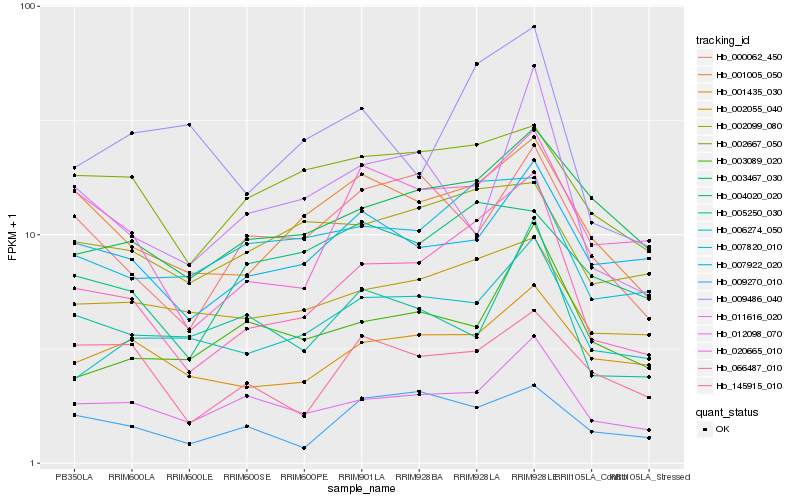
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_066487_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012098_070 |
0.1350491824 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 3 |
Hb_020665_010 |
0.138649458 |
- |
- |
PREDICTED: uncharacterized protein LOC103438977 isoform X2 [Malus domestica] |
| 4 |
Hb_001005_050 |
0.141201045 |
- |
- |
hypothetical protein Csa_3G599480 [Cucumis sativus] |
| 5 |
Hb_007820_010 |
0.1552461676 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: protein FLOWERING LOCUS D [Jatropha curcas] |
| 6 |
Hb_003467_030 |
0.1560845132 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 7 |
Hb_006274_050 |
0.1562309816 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 8 |
Hb_007922_020 |
0.1580058708 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 9 |
Hb_002055_040 |
0.159802533 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 10 |
Hb_009486_040 |
0.1617516569 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001435_030 |
0.1625200251 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 12 |
Hb_009270_010 |
0.1642741847 |
- |
- |
PREDICTED: uncharacterized protein LOC103421213 [Malus domestica] |
| 13 |
Hb_002667_050 |
0.1646324394 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 14 |
Hb_002099_080 |
0.1692514478 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 15 |
Hb_000062_450 |
0.169800572 |
- |
- |
hypothetical protein MTR_081s0009, partial [Medicago truncatula] |
| 16 |
Hb_003089_020 |
0.1733698607 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g18940, chloroplastic [Jatropha curcas] |
| 17 |
Hb_145915_010 |
0.1737426877 |
- |
- |
PREDICTED: uncharacterized protein LOC105630963 [Jatropha curcas] |
| 18 |
Hb_004020_020 |
0.1740959815 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g65820 [Jatropha curcas] |
| 19 |
Hb_005250_030 |
0.1745435873 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 20 |
Hb_011616_020 |
0.1750617728 |
- |
- |
hypothetical protein CICLE_v10004968mg [Citrus clementina] |