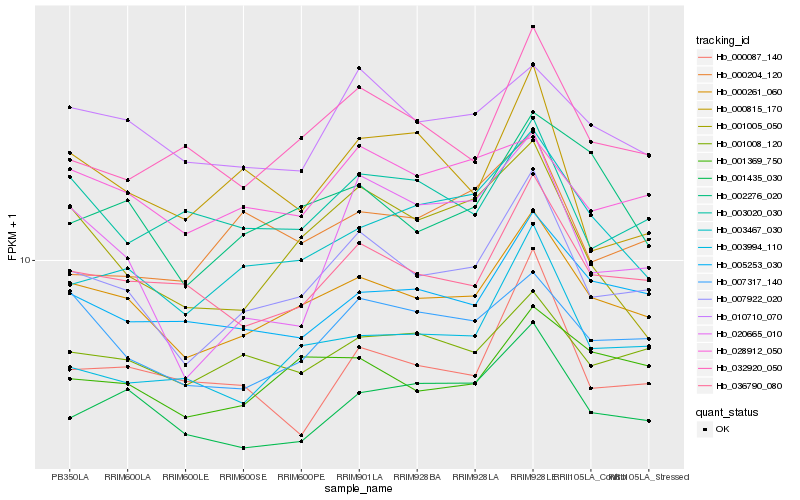
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007922_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 2 |
Hb_000261_060 |
0.0601608414 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 3 |
Hb_036790_080 |
0.0890594572 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 4 |
Hb_001435_030 |
0.0956242129 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 5 |
Hb_003994_110 |
0.0979257944 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 6 |
Hb_007317_140 |
0.0988677107 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 7 |
Hb_032920_050 |
0.100358283 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001008_120 |
0.1004463009 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 9 |
Hb_005253_030 |
0.1030279711 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_020665_010 |
0.1048242279 |
- |
- |
PREDICTED: uncharacterized protein LOC103438977 isoform X2 [Malus domestica] |
| 11 |
Hb_010710_070 |
0.1049181751 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_028912_050 |
0.106697876 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003020_030 |
0.1069437143 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_001369_750 |
0.1077450307 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 15 |
Hb_001005_050 |
0.10825476 |
- |
- |
hypothetical protein Csa_3G599480 [Cucumis sativus] |
| 16 |
Hb_003467_030 |
0.1084203397 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 17 |
Hb_000204_120 |
0.1086332389 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 18 |
Hb_000815_170 |
0.1092672686 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 19 |
Hb_000087_140 |
0.1095272238 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002276_020 |
0.1129064232 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |