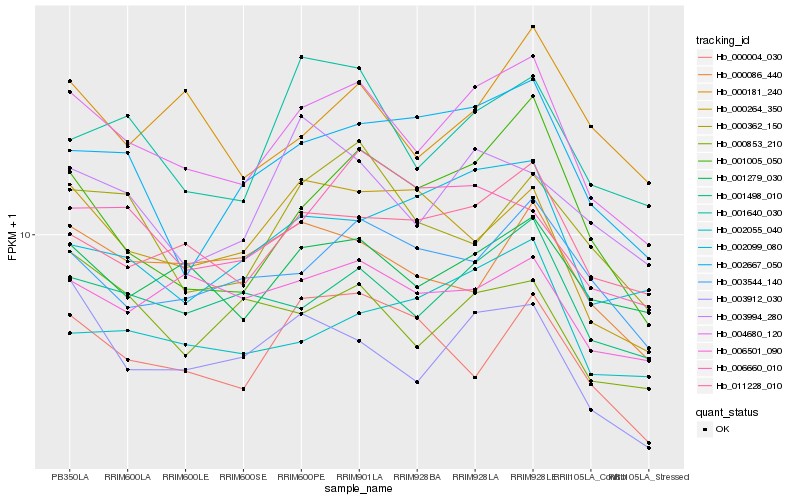
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004680_120 |
0.0 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 2 |
Hb_001498_010 |
0.0773408265 |
- |
- |
PREDICTED: uncharacterized protein LOC105648994 [Jatropha curcas] |
| 3 |
Hb_011228_010 |
0.0809260507 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_001279_030 |
0.082096299 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 5 |
Hb_003912_030 |
0.0861716269 |
- |
- |
PREDICTED: uncharacterized protein LOC105638204 [Jatropha curcas] |
| 6 |
Hb_001005_050 |
0.0910427736 |
- |
- |
hypothetical protein Csa_3G599480 [Cucumis sativus] |
| 7 |
Hb_003544_140 |
0.0936174583 |
- |
- |
PREDICTED: protein GDAP2 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_001640_030 |
0.0953130345 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 9 |
Hb_002667_050 |
0.0989180501 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 10 |
Hb_006501_090 |
0.0994561598 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 11 |
Hb_000853_210 |
0.0996615999 |
- |
- |
PREDICTED: uncharacterized protein LOC105642575 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002055_040 |
0.1032878837 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 13 |
Hb_003994_280 |
0.1041285806 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 14 |
Hb_000264_350 |
0.1042952861 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 15 |
Hb_000086_440 |
0.1045006514 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 16 |
Hb_002099_080 |
0.1051724513 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 17 |
Hb_006660_010 |
0.1051967342 |
- |
- |
PREDICTED: exocyst complex component EXO84C isoform X1 [Jatropha curcas] |
| 18 |
Hb_000362_150 |
0.1054007398 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 19 |
Hb_000181_240 |
0.1056217634 |
- |
- |
PREDICTED: uncharacterized protein LOC105637672 [Jatropha curcas] |
| 20 |
Hb_000004_030 |
0.1056459317 |
- |
- |
PREDICTED: Niemann-Pick C1 protein [Jatropha curcas] |