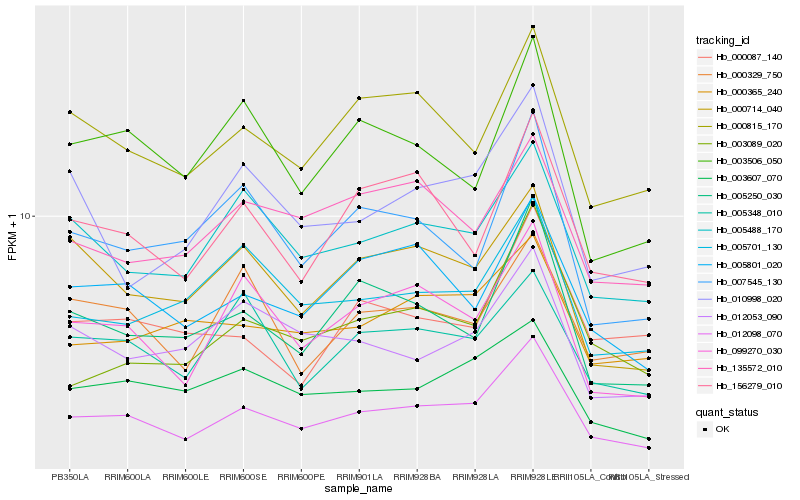
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012098_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 2 |
Hb_099270_030 |
0.0864097906 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 3 |
Hb_005488_170 |
0.0870663379 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 4 |
Hb_000815_170 |
0.088462049 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 5 |
Hb_003607_070 |
0.0893373029 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 6 |
Hb_005801_020 |
0.0903758982 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 7 |
Hb_005250_030 |
0.091918562 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 8 |
Hb_156279_010 |
0.0946505245 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000365_240 |
0.0947572625 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 10 |
Hb_005701_130 |
0.0995346716 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 11 |
Hb_005348_010 |
0.1004432444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003089_020 |
0.101634992 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g18940, chloroplastic [Jatropha curcas] |
| 13 |
Hb_012053_090 |
0.1047809113 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 14 |
Hb_135572_010 |
0.1047969256 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 15 |
Hb_000329_750 |
0.1051810372 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 16 |
Hb_000714_040 |
0.1053213061 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 17 |
Hb_010998_020 |
0.106955512 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 18 |
Hb_007545_130 |
0.1074313233 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000087_140 |
0.107704278 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003506_050 |
0.1090637992 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |