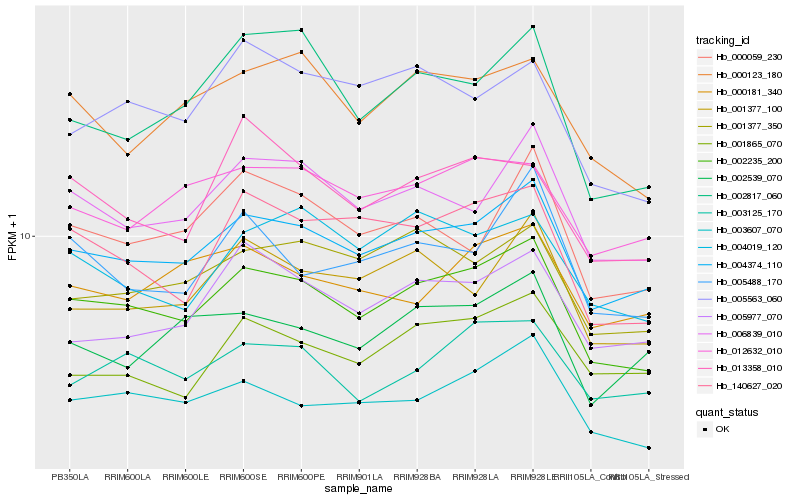
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_200 |
0.0 |
- |
- |
pattern formation protein, putative [Ricinus communis] |
| 2 |
Hb_004374_110 |
0.0458101556 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 3 |
Hb_003607_070 |
0.0720479713 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 4 |
Hb_006839_010 |
0.0768376319 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 5 |
Hb_140627_020 |
0.0786047016 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000123_180 |
0.0838327911 |
- |
- |
2-oxoglutarate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_003125_170 |
0.0864276726 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 8 |
Hb_001377_350 |
0.0877350075 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 9 |
Hb_001377_100 |
0.0884456776 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 10 |
Hb_012632_010 |
0.0902371745 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 11 |
Hb_005977_070 |
0.0922313784 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002817_060 |
0.0928488659 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 13 |
Hb_005563_060 |
0.0935286724 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 14 |
Hb_004019_120 |
0.0936542324 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 15 |
Hb_000059_230 |
0.0939440192 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 16 |
Hb_001865_070 |
0.0947192656 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000181_340 |
0.0953669115 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 18 |
Hb_005488_170 |
0.0955554049 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 19 |
Hb_013358_010 |
0.0962088021 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 20 |
Hb_002539_070 |
0.0974886895 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |