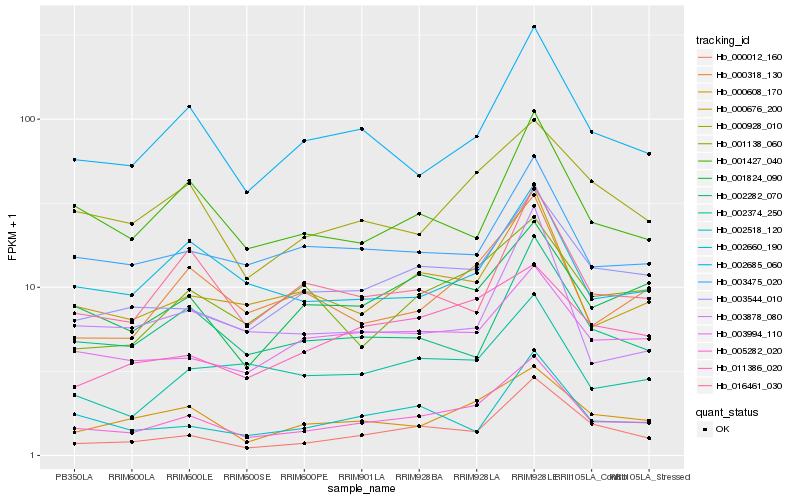
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005282_020 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |
| 2 |
Hb_003544_010 |
0.1004687172 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_000676_200 |
0.1073178373 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 4 |
Hb_000012_160 |
0.1085677459 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000608_170 |
0.1091434998 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 6 |
Hb_002685_060 |
0.1120589436 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 7 |
Hb_000318_130 |
0.1141300669 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000928_010 |
0.1175200132 |
- |
- |
Cell division protein ftsZ, putative [Ricinus communis] |
| 9 |
Hb_001824_090 |
0.1201915158 |
- |
- |
- |
| 10 |
Hb_003994_110 |
0.1217815834 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 11 |
Hb_003475_020 |
0.1234139684 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 12 |
Hb_002660_190 |
0.1283451005 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 13 |
Hb_011386_020 |
0.1303176156 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001427_040 |
0.1310356898 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 15 |
Hb_002282_070 |
0.1312953577 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 16 |
Hb_016461_030 |
0.1322069431 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_002374_250 |
0.1322660877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001138_060 |
0.1368715371 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 19 |
Hb_002518_120 |
0.1412543255 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |
| 20 |
Hb_003878_080 |
0.1437270675 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |