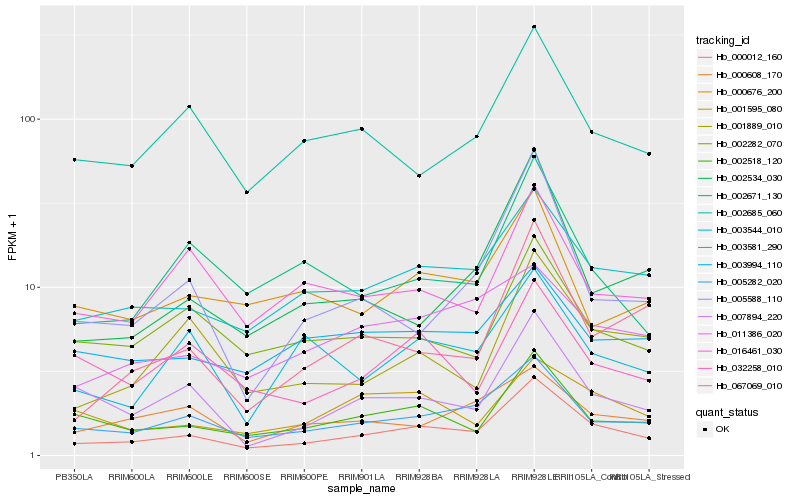
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005282_020 |
0.1085677459 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |
| 3 |
Hb_003544_010 |
0.1153086726 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 4 |
Hb_002518_120 |
0.1358034953 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |
| 5 |
Hb_007894_220 |
0.1446844485 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 6 |
Hb_002685_060 |
0.1536622015 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 7 |
Hb_002282_070 |
0.1566227653 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 8 |
Hb_001889_010 |
0.1581421655 |
transcription factor |
TF Family: Orphans |
PREDICTED: protein CHLOROPLAST IMPORT APPARATUS 2 [Jatropha curcas] |
| 9 |
Hb_002534_030 |
0.1609234178 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 10 |
Hb_002671_130 |
0.1627319131 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 11 |
Hb_067069_010 |
0.1631372649 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 12 |
Hb_005588_110 |
0.1641863623 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 13 |
Hb_016461_030 |
0.1645265704 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_011386_020 |
0.1657749899 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000676_200 |
0.166770498 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 16 |
Hb_032258_010 |
0.1676293356 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 17 |
Hb_003581_290 |
0.1678355632 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 3 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_003994_110 |
0.1684621731 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 19 |
Hb_000608_170 |
0.1687470118 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 20 |
Hb_001595_080 |
0.1694499835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |