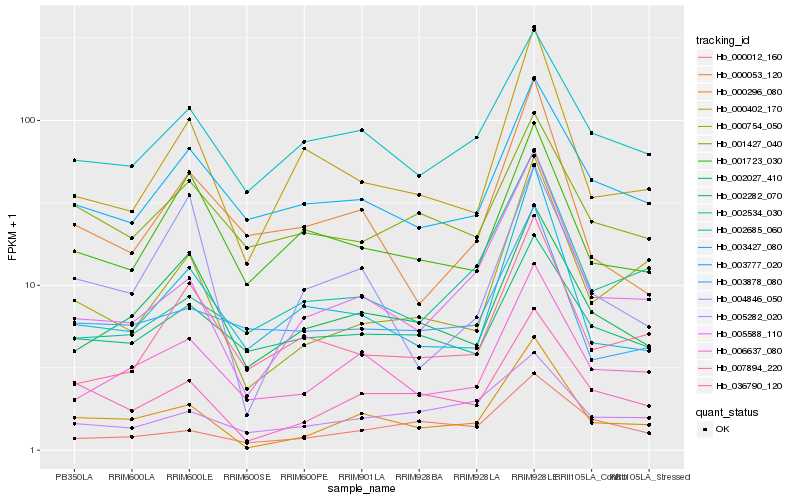
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000296_080 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35130 [Jatropha curcas] |
| 2 |
Hb_007894_220 |
0.1040380692 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 3 |
Hb_005588_110 |
0.1166828477 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 4 |
Hb_006637_080 |
0.1368862624 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000754_050 |
0.1391682866 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000402_170 |
0.1627029986 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 7 |
Hb_003427_080 |
0.1671362681 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002027_410 |
0.1696918729 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002685_060 |
0.1704563035 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 10 |
Hb_003777_020 |
0.1769061885 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 11 |
Hb_003878_080 |
0.1815599924 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |
| 12 |
Hb_000012_160 |
0.1844716737 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005282_020 |
0.1884666482 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |
| 14 |
Hb_000053_120 |
0.1889975632 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004846_050 |
0.1890636983 |
- |
- |
PREDICTED: uncharacterized protein ycf23-like [Jatropha curcas] |
| 16 |
Hb_001723_030 |
0.1902510537 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002282_070 |
0.1904723532 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 18 |
Hb_036790_120 |
0.1906576824 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002534_030 |
0.1921780302 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 20 |
Hb_001427_040 |
0.1922489004 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |