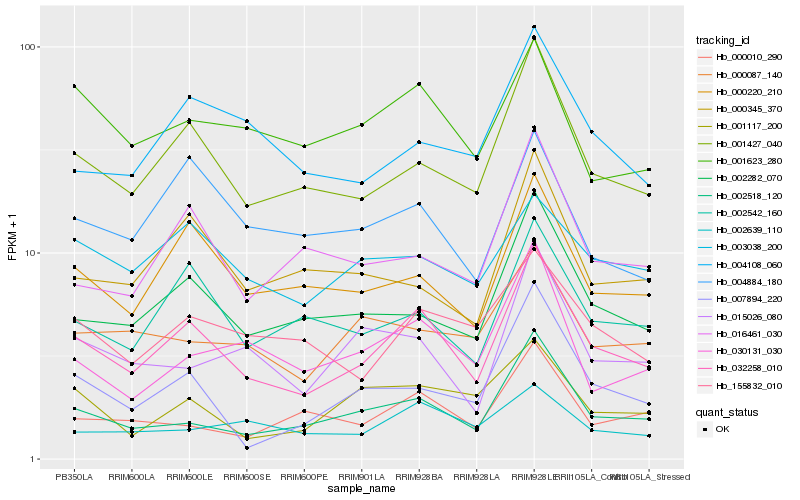
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032258_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 2 |
Hb_001427_040 |
0.1004341848 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 3 |
Hb_002518_120 |
0.1157233288 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |
| 4 |
Hb_002282_070 |
0.1231691553 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 5 |
Hb_000220_210 |
0.1247870196 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002542_160 |
0.1297476625 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_155832_010 |
0.1426279884 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 8 |
Hb_016461_030 |
0.1455031374 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_007894_220 |
0.1466192052 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 10 |
Hb_004108_060 |
0.1472880302 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 11 |
Hb_030131_030 |
0.1481123692 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g42310, mitochondrial [Jatropha curcas] |
| 12 |
Hb_004884_180 |
0.1496110294 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000010_290 |
0.1506769324 |
- |
- |
- |
| 14 |
Hb_015026_080 |
0.1507877772 |
- |
- |
- |
| 15 |
Hb_000345_370 |
0.1509588682 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 16 |
Hb_001623_280 |
0.1523079666 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000087_140 |
0.1528010417 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001117_200 |
0.1535061642 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002639_110 |
0.1557209124 |
- |
- |
hypothetical protein L484_011673 [Morus notabilis] |
| 20 |
Hb_003038_200 |
0.1560163047 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |