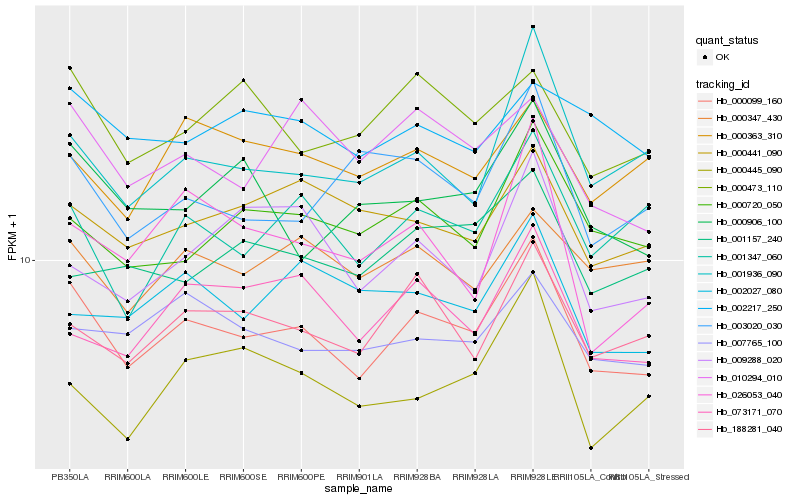
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_160 |
0.0 |
- |
- |
PREDICTED: potassium transporter 3 isoform X2 [Populus euphratica] |
| 2 |
Hb_188281_040 |
0.0798287665 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 3 |
Hb_001936_090 |
0.0835346887 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000720_050 |
0.0842611349 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001347_060 |
0.0847959064 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_010294_010 |
0.0872858739 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 7 |
Hb_007765_100 |
0.0877343545 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 8 |
Hb_000906_100 |
0.0883455285 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |
| 9 |
Hb_000441_090 |
0.0884640394 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 10 |
Hb_000347_430 |
0.0893890076 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 11 |
Hb_073171_070 |
0.0900268203 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 12 |
Hb_000473_110 |
0.0910856753 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 13 |
Hb_000363_310 |
0.0933502588 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 14 |
Hb_003020_030 |
0.0949743158 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_002027_080 |
0.0954743099 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 16 |
Hb_000445_090 |
0.0955302264 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 17 |
Hb_002217_250 |
0.0963047054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_009288_020 |
0.0972796921 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 19 |
Hb_026053_040 |
0.0979718939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001157_240 |
0.0979767765 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |