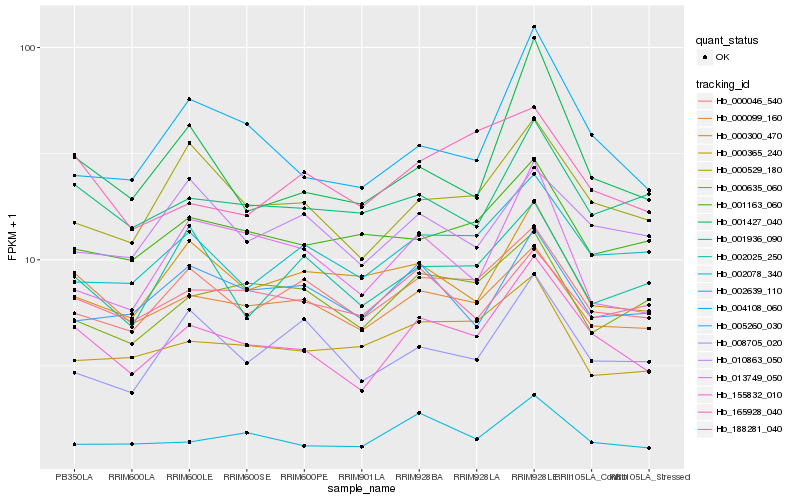
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155832_010 |
0.0 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 2 |
Hb_000529_180 |
0.1038293616 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000099_160 |
0.1061625972 |
- |
- |
PREDICTED: potassium transporter 3 isoform X2 [Populus euphratica] |
| 4 |
Hb_000046_540 |
0.1068953015 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 5 |
Hb_004108_060 |
0.1128416955 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 6 |
Hb_002639_110 |
0.1141013831 |
- |
- |
hypothetical protein L484_011673 [Morus notabilis] |
| 7 |
Hb_002078_340 |
0.1157219872 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001936_090 |
0.1163224168 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001427_040 |
0.1229391535 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 10 |
Hb_165928_040 |
0.127463824 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase component HRD3A [Jatropha curcas] |
| 11 |
Hb_000635_060 |
0.1277802571 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 12 |
Hb_000365_240 |
0.1281980265 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 13 |
Hb_005260_030 |
0.1286732614 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 14 |
Hb_001163_060 |
0.128721222 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 15 |
Hb_013749_050 |
0.1289471399 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000300_470 |
0.1292120287 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 17 |
Hb_188281_040 |
0.1299506731 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 18 |
Hb_002025_250 |
0.1307776835 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 19 |
Hb_010863_050 |
0.1308245517 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 20 |
Hb_008705_020 |
0.1308951251 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |